Figure 1.
PHOX2A Overexpression Drives the Generation of Bona Fide Oculomotor Neurons from Stem Cells
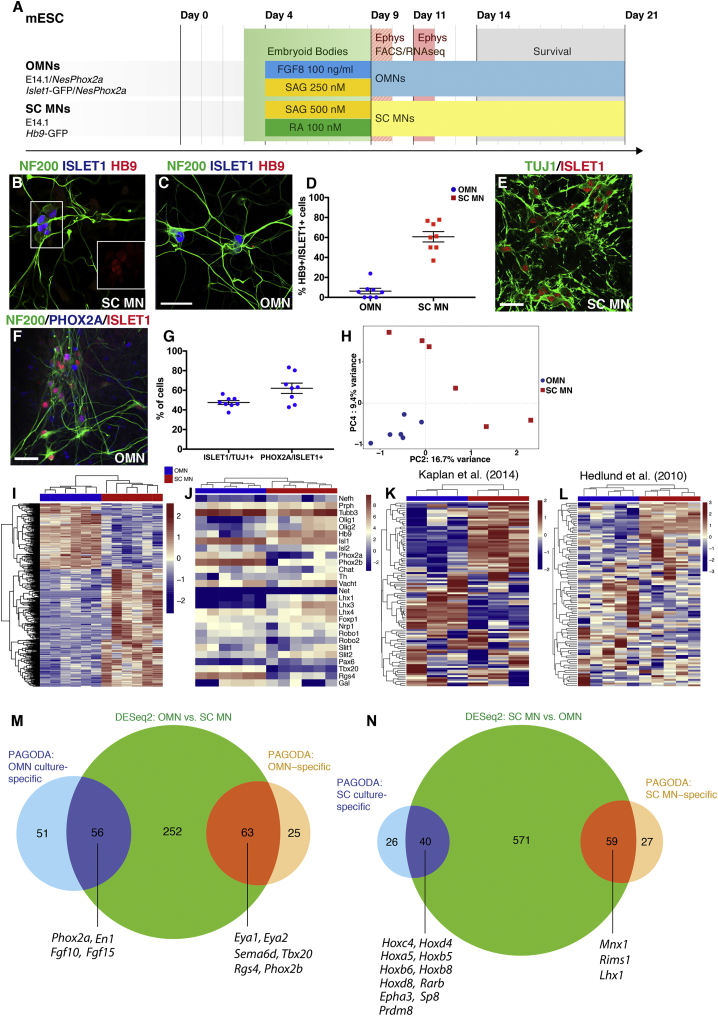
(A) Time line depicting in vitro protocols for the generation of resistant oculomotor neuron (OMN) and vulnerable spinal cord motor neuron (SC MN). The mESC column reports the four cell lines used in this study. Patterning was performed for 5 days following EB formation. mRNA-seq coupled with FACS was performed on EB dissociation day (day 9 of the protocol). The survival assay was performed 5 days after EB dissociation (day 14 to 21).
(B and C) SC MNs generated from E14.1 ESCs express HB9, ISLET1, and NF200 (B). OMNs generated from E14.1 ESCs overexpressing the transcription factor Phox2a under the Nestin enhancer co-express ISLET1 and NF200 in the absence of HB9 (C). Scale bar, 60 μm.
(D) Percentage of HB9+/ISLET1+ cells in SC MN (64.1% ± 5.2%, mean ± SEM, n = 4) and OMN (6.3% ± 1.3%, mean ± SEM, n = 4) cultures, respectively (at least 120 Islet1+ cells counted per condition, bar and whiskers represent means ± SEM).
(E) Microphotographs showing ISLET1+/TUJ1+ cells in OMN cultures.
(F) OMNs also express the specific marker PHOX2A as indicated by asterisks. Scale bar, 100 μm (applies also to E).
(G) Quantification of ISLET1+ over TUJ1+ cells demonstrates that half the neuronal population appears to be ISLET1+ (47.5% ± 5.9%, mean ± SEM, n = 4; total number of TUJ1+ cells counted: 1,325). Quantification of PHOX2A+ over ISLET1+ cells (experiments performed in quadruplicates with two technical replicates each, total number of Islet1+ cells counted: 746) indicates that 62% ± 5.2% (mean ± SEM, n = 4) of the ISLET1+ population is also PHOX2A+. All quantifications were performed 5 days after EB dissociation, and experiments were conducted in quadruplicates including two technical replicates per experiment (bar and whiskers represent means ± SEM).
(H) PCA of OMN and SC MN samples based on all genes expressed confirmed cell differential identities. mRNA-seq analysis of OMNs and SC MNs isolated by FACS was performed after 5 days of patterning (day 9 of the protocol).
(I) A total of 1,017 DEGs was found between the two different cell types (adjusted p < 0.05, n = 6). Heatmap shows the top 500 most significant DEGs by adjusted p value.
(J–L) Heatmap of expected progenitors and motor neuron transcripts but also OMN and SC MN specific transcripts obtained in the two generated motor neuron populations (J and L). Using the top 100 DEGs obtained from our RNA-seq analysis (K), we could separate datasets originating from in vivo microarray studies on early postnatal (J) and adult (L) rodent OMNs and SC MNs.
(M and N) Venn diagrams showing gene sets enriched either in brain stem cultures or OMN-specific as revealed by PAGODA analysis (M), and gene sets preferentially found in spinal cord cultures or MN specific (N).