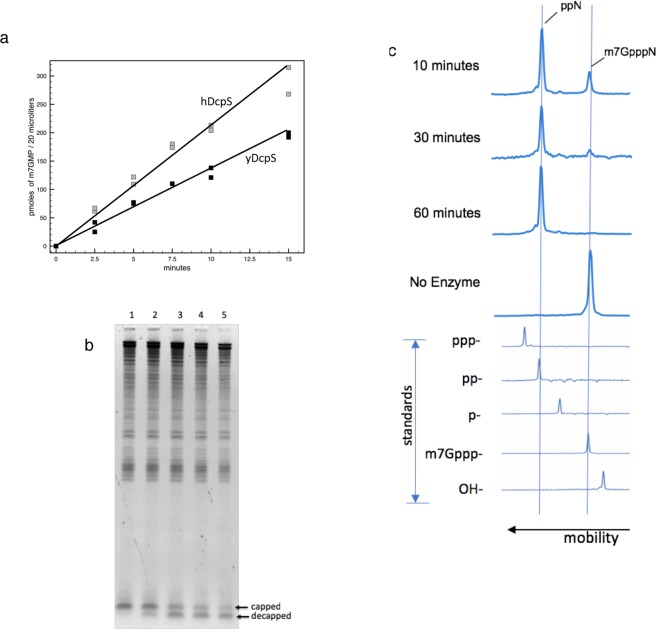
Figure 2.
Biochemical Assays for DcpS. (a) Conversion of m7GpppA dinucleotide to m7GMP. A 200 µL reaction in decapping buffer containing 50 µM m7GpppA and either 25 nM yDcpS (black □) or 42 nM hDcpS (grey □) was incubated at 37 °C. 20 µL samples were withdrawn at the indicated times. The reactions were terminated by incubation at 70 °C. Picomoles of m7GMP product were determined by LC-MS. (b) Decapping of a 25mer Cap 1 RNA by yDcpS. A 30 µL reaction containing 300 ng of total E. coli RNA and 60 ng of 25mer Cap 1 RNA was incubated for 3 hours at 37 °C with 130 ng of yDcpS in 10 mM MES pH 6.5 and 1 mM EDTA. At 0 minutes a 5 µL aliquot was mixed with 2X RNA loading dye stop solution (Lane 1). Likewise 5 µL aliquots were taken at 5, 60, 120, and 180 min (Lanes 2 to 5, respectively). The RNA aliquots were analyzed by 15% TBE-Urea PAGE stained with SYBR Gold. (c) Capillary electrophoresis of 3′-FAM-labeled 5′-capped 25mer RNA. Top panel shows a representative example of the decapping reaction progress. The 25mer Cap 0 RNA was incubated with yDcpS for various times indicated on the left. Bottom panel shows the mobility shift for standards of 25mer RNAs containing different 5′ ends. All data were plotted relative to GeneScanTM120 LIZTM Applied Biosystems standards.