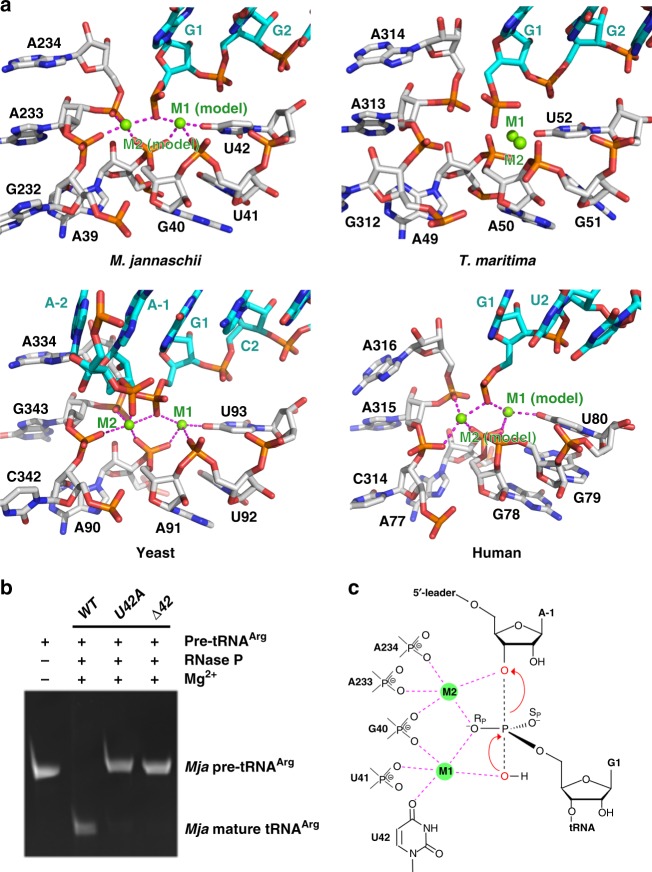
Fig. 7.
The catalytic center of MjaRNase P. a Close-up views of the catalytic centers of MjaRNase P (top left), T. maritima RNase P (top right, PDB: 3Q1Q), yeast RNase P (bottom left, PDB: 6AH3), and human RNase P (bottom right, PDB: 6AHU). Two catalytic Mg2+ ions (M1 and M2) in the active site of MjaRNase P and human RNase P shown in green spheres were modeled based on the yeast RNase P-tRNA complex structure. RPR and tRNA are showed in stick representation and colored in silver and cyan, respectively. The coordination of the two Mg2+ ions is denoted by magenta dashed lines. b In vitro pre-tRNA processing assay of the MjaRNase P holoenzyme reconstituted with WT, U42A and ΔU42 RPR, respectively. c Proposed reaction mechanism for 5′-leader cleavage of pre-tRNA by MjaRNase P. The reactive oxygens are colored in red, the pre-tRNA scissile phosphate is depicted in a transition state, and the interactions between catalytically important nucleotides and reactive oxygens mediated by Mg2+ ions (M1 and M2) are shown as magenta dashed lines