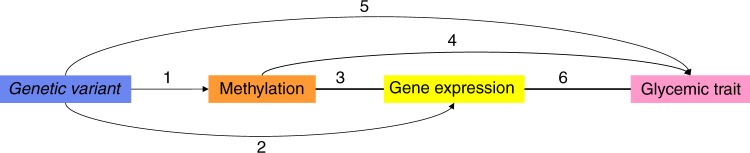
Fig. 1.
Overview of the cross-omics analysis. (1) Methylation quantitative trait loci (meQTL). (2) Expression quantitative trait loci (eQTL). (3) Expression quantitative trait methylation (eQTM). (4) Epigenome-wide association study (EWAS) and Mendelian randomization (MR). (5) Genome-wide association study (GWAS). (6) The association of gene expression expressed in the glucose or insulin metabolism-related tissues and glycemic traits. Results in 1, 2, 3 were extracted from the summary statistics from Biobank-based Integrative Omics Study (BIOS) database (n = 3814). Results in 4 was the results in the current EWAS (discovery phase, n = 4808, replication phase, n = 11,750) and the two-sample Mendelian randomization based on the BIOS database (n = 3814) and GWAS results of Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC). Results in 5 was from the GWAS results of MAGIC or the DIAbetes Genetics Replication And Meta-analysis consortium (DIAGRAM, n = 96,496–452,244). Results in 6 was based on the summary statistics of Genotype-Tissue Expression project (GTEx) and MAGIC or DIAGRAM (n = 153–491)