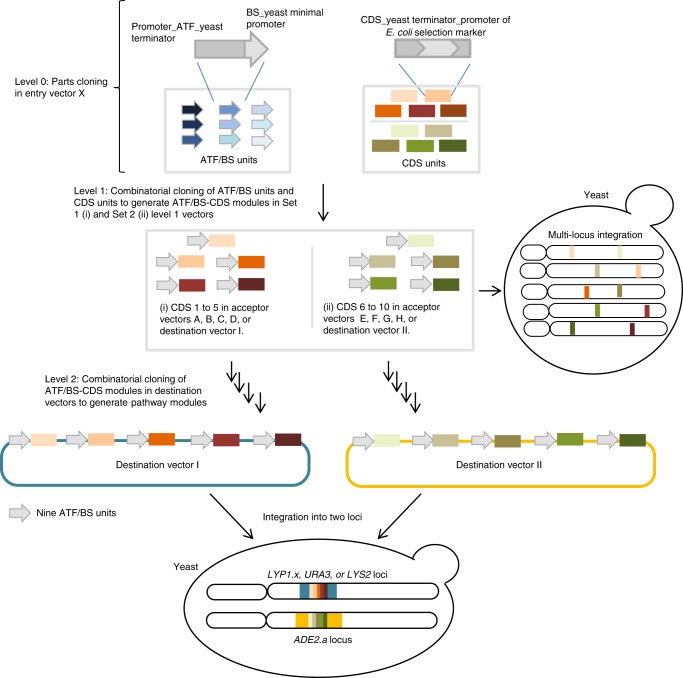
Fig. 1.
The COMPASS workflow. The workflow encompasses three assembly levels. At Level 0, nine ATF/BS units are assembled into Entry vector X. Each ATF/BS unit harbors an inducible promoter to drive ATF expression, the ATF CDS, a yeast terminator, and promoter fragments containing one or more copy(s) of the ATF's BS within a minimal CYC1 promoter. In separate reactions, each of the ten CDSs is combined with a yeast terminator and the promoter of an E. coli selection marker (which defines the vector of the next assembly level) into Entry vector X to generate CDS units. At Level 1, two parallel combinatorial clonings are performed: (i) The nine different ATF/BS units (gray arrows), five CDS units (colored rectangles), and a set of linearized auxotrophic marker vectors (Acceptor vectors A–D, and Destination vector I) are subjected to overlap-based cloning in a single reaction tube (using NEBuilder HiFi). (ii) Similarly, nine ATF/BSs, five other CDSs, and a set of dominant-marker vectors (Acceptor vectors E–H, and Destination vector II) are combined. Using this approach, COMPASS generates ten groups of vectors, each containing nine plasmids (90 plasmids in total). Thereafter, the ten groups of ATF/BS-CDS modules of Level 1 may be integrated into ten defined loci of the yeast genome. At Level 2, ten libraries of ATF/BS-CDS modules are established in Destination vector I and Destination vector II (five libraries in each vector). The Destination vectors I are integrated into the LYS2, URA3, (or LYP1.x) loci, while Destination vectors II are integrated into the ADE2.a locus