Fig. 3.
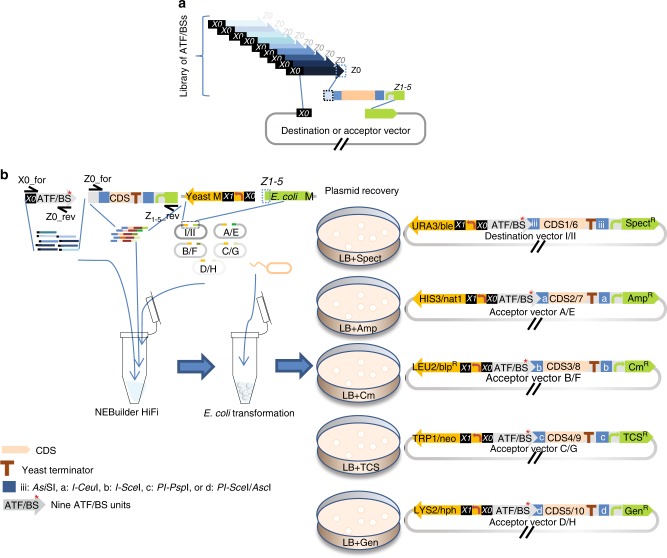
Combinatorial assembly of ATF/BS and gene units in the modules. a Combinatorial assembly of ATF/BS units upstream of CDS units. Nine ATF/BS units and CDS units from the Entry vectors X are PCR-amplified. The 5′ regions of ATF/BS units overlap with the X0 sequence of the vector backbone, while their 3′ regions are identical to the 30 bp of the 3′ end of the minimal CYC1 promoter (Z0) and overlap with the forward primer amplifying the CDS units. The 3′ regions of the CDS units overlap with Z1–Z5 of five linearized vectors. b The COMPASS workflow to generate ATF/BS-CDS modules in vectors of either Set 1 or Set 2 (1/2). The PCR-amplified ATF/BS unites (primers X0_for/Z0_rev, on Entry vectors X-ATF/BS), five CDS units (primers Z0_for/Z1_rev/Z2_rev/Z3_rev/Z4_rev/Z5_rev at molar ratio 5:1:1:1:1:1, on mixed Entry vectors X-CDS) and five linearized Destination vectors I/II and Acceptor vectors A/E, B/F, C/G, and D/H are mixed at molar ratio 2:2:1 for in vitro overlap-based cloning in a single tube to generate different ATF/BS-CDS modules in the diverse vectors by providing the missing promoter sequences of the E. coli markers within the assembly units. Therefore, libraries of Destination vectors I-CDS1/II-CDS6, Acceptor vectors A-CDS2/E-CDS7, B-CDS3/F-CDS8, C-CDS4/G-CDS9, or D-CDS5/H-CDS10 are generated. X0_for overlaps with X0, Z0_rev overlaps with Z0_for, while primers Z1_rev–Z5_rev overlap with the downstream (right) HR of the linearized vector (called Z1–Z5). Light-orange arrow, CDS. Brown “T”, yeast terminator. Blue squares, iii: AsiSI, a: I-CeuI, b: I-SceI, c: PI-PspI, d: PI-SceI / AscI. Gray arrows, nine ATF/BS units. For simplicity, IPTG-inducible promoters and terminators are not included in the figure. X0, Z0–Z5 are explained in footnote to Supplementary Data 4