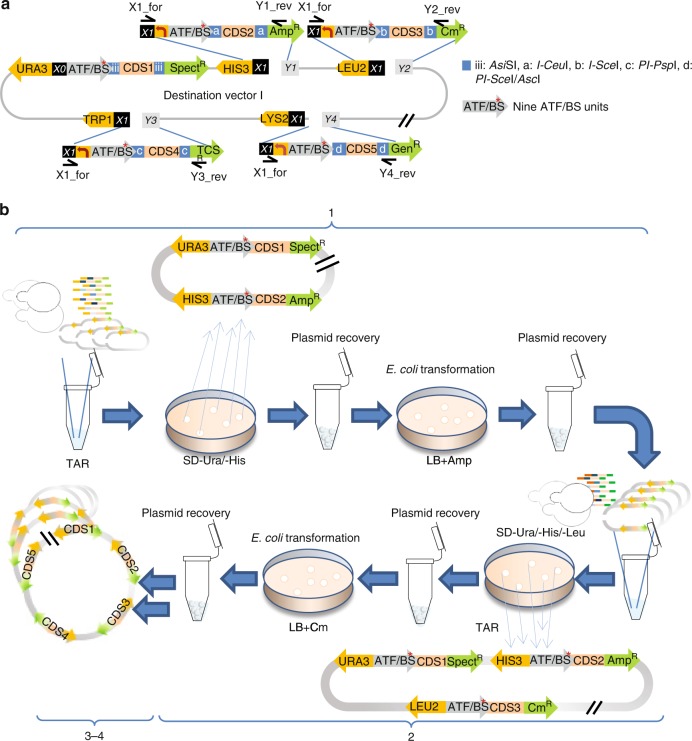
Fig. 4.
Combinatorial cloning of pathway modules into Destination vector I. a Combinatorial cloning of pathway genes into Destination vector I. The libraries of PCR-amplified (i) ProHIS3-ATF/BS-CDS2-ProAmpR-AmpR-TerAmpR (primers X1_for/Y1_rev, on Acceptor vector A), (ii) ProLEU2-ATF/BS-CDS3-ProCmR-CmR-TerCmR (primers X1_for/Y2_rev, on Acceptor vector B), (iii) ProTRP1-ATF/BS-CDS4-ProTCSR-TCSR-TerTCSR (primers X1_for/Y3_rev, on Acceptor vector C), and (iv) ProLYS2-ATF/BS-CDS5-ProGenR-GenR-TerGenR (primers X1_for/Y4_rev, on Acceptor vector D) modules are successively assembled in sites p2–p5, correspondingly, starting with the Destination vectors I-CDS1 library (see Fig. 3), in four rounds of combinatorial cloning. b The COMPASS workflow for combinatorial assembly of ATF/BS and gene modules into Destination vector I. The mixed ATF/BS-CDS2 modules are assembled using TAR in site p2 of Destination vectors I-CDS1. Yeast cells with successful constructs grow on SC-Ura/-His medium. Cells are scraped from the plates, the plasmid library is extracted to obtain a pool of all randomized members, transformed into E. coli, and cells are grown on LB plates containing ampicillin. Cells are scraped from the plates to extract the plasmid library (1). The ATF/BS-CDS3 modules are assembled in site p3 of the Destination vectors I-CDS1-CDS2 library. Yeast cells with successful constructs grow on SC-Ura/-His/-Leu medium (2). The libraries of ATF/BS-CDS4 and ATF/BS-CDS5 modules are cloned into sites p4 and p5, respectively (3 and 4). Gray arrows, nine ATF/BS units. Blue squares, iii: AsiSI, a: I-CeuI, b: I-SceI, c: PI-PspI, d: PI-SceI / AscI. For simplicity, the IPTG-inducible promoters and terminators are not included in the figure. X0 and X1 are explained in footnote to Supplementary Data 4. Y1–Y4 overlap with the last 30 bp of terminators of the AmpR, CmrR, TCSR, and GenR genes