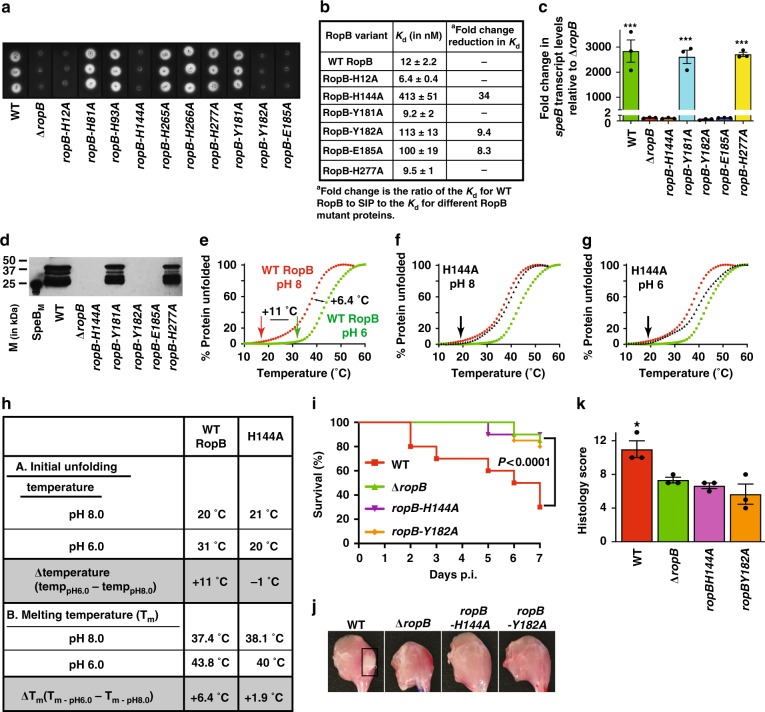
Fig. 6.
The histidine switch in RopB is critical for speB expression and GAS virulence. a SpeB protease activity made by each isogenic ropB mutant strain. b RopB–SIP-binding constants for the indicated recombinant RopB mutant proteins. Characterization of ropB mutant strains for speB transcript levels (c), and secreted SpeB levels (d). P values (***P < 0.001) of the indicated strains were compared to ∆ropB mutant strain. P values were determined by t-test. Source data are provided as a Source Data file. e Thermal stability of WT RopB was determined by a thermofluor assay. Thermal shift assay results for WT RopB in below (pH 6, colored green) and above neutral pH (pH 8, colored red) are shown. The temperature at which initial unfolding of WT RopB occurs are marked by vertical arrows and color coded. The horizontal two-headed arrows indicate the differences in melting temperatures (Tm) and in temperatures at which initial unfolding occurs between below and above neutral pH. The thermal stability curves of H144A mutant protein in above (pH 8) (f) and below (pH 6) (g) neutral pH are overlaid onto the thermal stability curves of WT RopB and colored in black. The temperature at which initial unfolding of H144A mutant protein occurs is marked by black vertical arrows. h Comparison of thermal shift assay results of WT RopB and H144A mutant protein. i Twenty outbred CD-1 mice per strain were injected intramuscularly with 1 × 107 CFUs of each indicated bacterial strain. Kaplan–Meier survival curve with P values derived by log rank-test are shown. j Analyses of gross hindlimb lesions from mice infected with each indicated strain. Analysis of the infected hindlimbs was performed at 48 h postinoculation. Larger lesion with extensive tissue damage in WT-infected mice in pH 6 is boxed (black box). k Histopathology scores of hindlimb lesions from mice infected with each indicated strain. Histopathology analysis of the infected hindlimbs was performed at 48 h postinoculation. Data are expressed as means + standard deviation. P values (*P < 0.05) relative to ∆ropB mutant strain are shown. P values were determined by t test