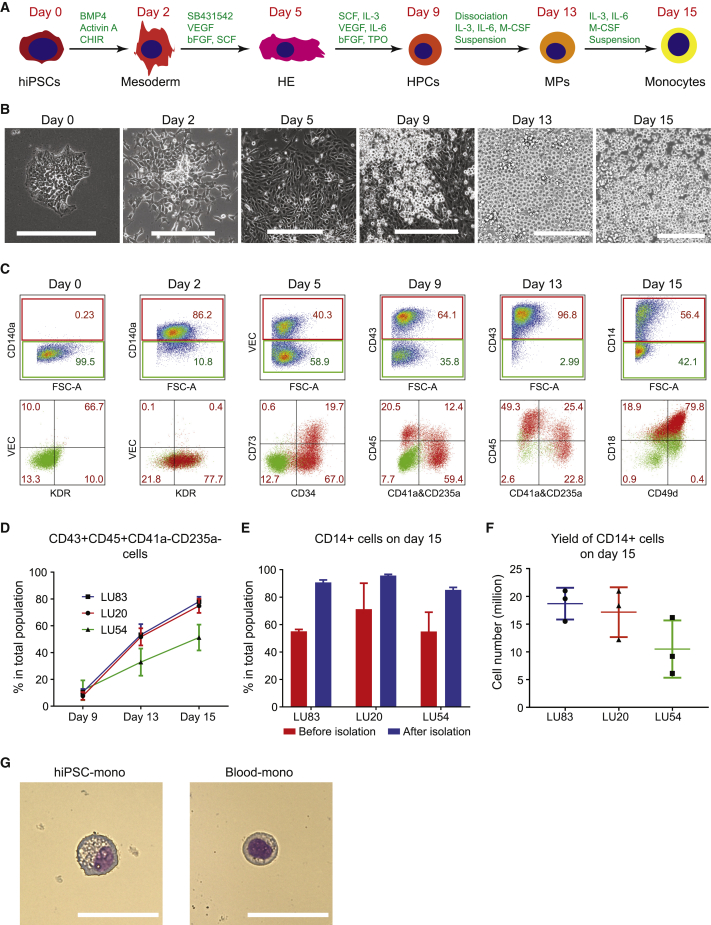
Figure 1.
Differentiation of CD14+ Monocytes from hiPSCs
(A) Schematic overview of CD14+ monocyte differentiation protocol from hiPSCs.
(B) Bright-field images of representative cellular morphology at day 0 (undifferentiated hiPSCs), day 2 (mesoderm), day 5 (HE), day 9 (HPCs), day 13 (MPs), and day 15 (CD14+ monocytes). Scale bar represents 200 μm.
(C) FACS analysis of stage-specific markers at day 0, day 2, day 5, day 9, day 13, and day 15 of differentiation from a representative hiPSC line (LU83). Positive populations are gated in the upper panels, and their percentages are shown in red in both upper and lower panels.
(D) Quantification of the percentage of myeloid lineage cells (CD43+CD45+CD41a−CD235a−) in the total cell population at day 9, day 13, and day 15 of differentiation. Quantification of three independent experiments from three hiPSC lines (LU83, LU20, and LU54) is shown.
(E) Quantification of the percentage of CD14+ cells at day 15 of differentiation before and after isolation using CD14+ MACS. Quantification of three independent experiments from three hiPSC lines (LU83, LU20, and LU54) is shown.
(F) Yield of CD14+ monocytes at day 15 of differentiation from three hiPSC lines and three independent experiments. Yield of monocytes is equal to the total cell number multiplied by percentages of CD14+ cells.
(G) Giemsa staining of hiPSC-mono isolated at day 15 of differentiation from one representative hiPSC line (LU83) and Blood-mono. Scale bar represents 50 μm.
Error bars are ±SD of three independent experiments in (D–F). See also Figure S1 and Videos S1 and S2.