Abstract
Myocardial infarction (MI) is primarily caused by ischemic heart or coronary artery disease and is a major cause of mortality worldwide. Thus, it is necessary to establish reliable biochemical markers for the early diagnosis of MI. MicroRNAs (miRNAs or miR) have been demonstrated to circulate in biological fluids and are enclosed in extracellular vesicles, including exosomes. The current study assessed the differential expression of exosomal miRNAs in the plasma of patients with MI and healthy individuals, and the possible mechanism involved. Plasma-derived exosomes were isolated from patients with MI and healthy control individuals, and vesicles with a membrane were observed using transmission electron microscopy. Furthermore, an exosomal miRNA expression profile was compared between patients with MI and healthy individuals using a miRNA microarray. Significantly differentially expressed miRNAs were validated using reverse transcription-quantitative polymerase chain reaction. To the best of our knowledge, the present study was the first to demonstrate that miR-183 was markedly upregulated in patients with MI compared with healthy individuals. In addition, the relative exosomal miR-183 level increased with the degree of myocardial ischemic injury. Additionally, GO and KEGG analyses demonstrated that miR-183 is primarily involved in cell communication, protein kinase activity regulation and adrenergic signaling in cardiomyocytes. Furthermore, a protein-protein interaction network of all the miR-183 target genes was constructed. The results demonstrated that five core genes (PPP2CB, PPP2CA, PRKCA, PPP2CA, PPP2R5C and PPP2R2A) in the PPI network were also associated with protein kinase activity regulation and adrenergic signaling in cardiomyocytes. Taken together, these data demonstrate that exosomal miR-183 derived from the serum of patients with MI may be a novel diagnostic biomarker involved in the regulation of protein kinase activity. miR-183 may therefore be further developed for clinical use to benefit patients with coronary artery diseases.
Keywords: myocardial ischemic injury, exosome, microRNA-183, protein kinase activity
Introduction
Myocardial infarction (MI) has a high morbidity and mortality worldwide (1). Percutaneous coronary intervention and thrombolytic therapy are widely accepted as standard treatments for MI and the mortality rate of patients decreases gradually in the acute phase (2). However, the mortality rate of coronary artery disease remains high in the general population (3). Thus, it is necessary to identify novel biomarkers for the early diagnosis of myocardial ischemic injury, which may provide valuable benefits to current therapies.
Certain cyclic myocardial injury markers, including high-sensitivity cardiac troponin (4), natriuretic peptide (5) and cyclic microRNA (miRNA) (6) have garnered a great deal of attention. However, the number of available markers remains limited. Exosomes are small cell-derived vesicles, with a diameter of 30–120 nm, which are located in many, if not all biological fluids (7). These contain mRNA, miRNAs, long non-coding RNAs, DNA, lipids and other small molecules (7). miRNAs have been studied in many diseases due to their utility in disease diagnoses, to monitor therapy and to predict the probability of disease recurrence; a recent study demonstrated that various exosomal miRNA biomarkers may be useful for the diagnosis of various diseases, including nervous system diseases, malignant tumors and respiratory diseases (8). For example, miR-21, miR-26a and miR-146a were expressed in bronchoalveolar lavage exosomes, however the expression of other miRNAs involved in the initiation and progression of inflammation in sarcoidosis was not observed (8). In addition, circulating muscle-specific miRNAs were different between COPD and non-COPD subjects (8). The aforementioned study suggested that the plasma level of exosomal miRNA can reflect changes within the skeletal muscle and so can act as a biomarker of skeletal muscle dysfunction. In fact, exosomal miRNAs have been considered a noninvasive diagnostic biomarkers and therapeutic target in respiratory diseases (8). However, the diagnostic value of exosomal miRNAs remains unknown in patients with myocardial ischemic injury. Thus, there is a growing need for diagnostic markers to improve the outcome of patients with myocardial ischemic injury.
To the best of our knowledge, the current study is the first to compare the miRNA profile of dysregulated plasma exosomes in patients with MI compared with healthy controls. Furthermore, the present study assessed the possible mechanism that underlies the regulation of miRNA in myocardial ischemic injury.
Materials and methods
Collection and preparation of plasma samples
Plasma samples from 14 patients with MI, 10 patients with stable angina pectoris and 14 healthy individuals were collected between January 2016 and December 2016 from the First Hospital of Shanxi Medical University (Taiyuan, China). The current study recruited 20 males and 18 females, aged 41–78 years. The mean age of these subjects was 47.6 years. Following collection, the samples were stored at −80°C. These patients were diagnosed using coronary angiograms, electrocardiograms and cardiac troponin tests, and in accordance with previously published diagnostic criteria guidelines (9,10). The exclusion criteria were as follows: i) Patients unable to co-ordinate with doctors, ii) patients with multiple medical complications or iii) patients who received drugs treatment associated with angiocardiopathy within one month of recruitment. Written informed consent was obtained from all the participants and the present study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (Taiyuan, China).
Exosomes and exosomal RNA isolation
Plasma from each participant was filtered using a Millipore Millex-AA syringe (cat. no. SLAA033SB; Merck KGaA, Darmstadt, Germany) prior to isolation. An exoEasy Maxi kit (Qiagen GmbH, Hilden, Germany) was used to isolate exosomes from 4 ml of pre-filtered plasma according to the manufacturer's protocol. Following exosomal isolation, the remaining plasma was diluted with PBS (1:1) and utilized as a control. For exosomal RNA extraction, an exoRNeasy Serum/Plasma Maxi kit (Qiagen GmbH) was used to isolate total exosome RNA from 4 ml pre-filtered plasma according to the manufacturer's protocol. Finally, an RNeasy MinElute spin column was placed into a 1.5 ml collection tube and 14 µl of RNase-free water (both Qiagen GmbH) was directly added to the center of the spin column membrane. The lid was closed gently, the column was left to stand for 1 min and was then centrifuged for 1 min at 5,000 × g and 25°C to elute the RNA. Collected RNA was then used in the subsequent experiments.
Transmission electron microscopy
Following exosome isolation, extracellular vesicles were fixed with 4% paraformaldehyde for 30 min at 25°C and treated as previously described (11). The morphological characteristics of plasma-derived extracellular vesicles were observed via transmission electron microscopy at a magnification of ×30,000 (11).
Microarray analysis
miRNA microarray analysis was performed using the Human miRNA OneArray® v6 (PhalanxBio, Inc., San Diego, CA, USA). This contained triplicate 2539 unique miRNA probes from humans (miRBase release v20; http://www.mirbase.org/), each printed in technical triplicate, with 114 experimental control probes. The experiment was performed according to a previous study (12), and fold-change (FC) was calculated. FC=log2avg (MI)-log2avg (healthy control). The heatmap of differentially expressed miRNAs was constructed by R software (version 3.4.2; Vienna University of Economics and Business, Vienna, Austria).
RNA preparation and reverse transcription-quantitative polymerase chain reaction
Total RNA was isolated using the Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol and RNA (0.5 µg) was utilized to synthesize cDNA using Super Scriptase III (Invitrogen; Thermo Fisher Scientific, Inc.) with stem-loop RT primers (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTGAA-3′). The resultant cDNA was used for PCR. The thermocycling conditions were as follows: Denaturation at 95°C for 3 min, then 40 cycles of 95°C for 15 sec, 58°C for 30 sec and 72°C for 30 sec. To detect miR-183, the polymerase a method was utilized, followed by SYBR-Green qPCR (Thermo Fisher Scientific, Inc.) as described previously (3). Relative gene expression data was produced using the 2−∆∆Cq method (13). U6 was used as an internal control. The U6 primers used in the qPCR analysis were as follows: Forward: 5′-GCACTTTATTGGTCCATCATCC-3′; reserve: 5′-GACACCTGGCTCTTTTTGATTC-3′. The miR-183 primers used in the qPCR analysis were as follows: Forward: 5′-CGCGGTATGGCACTGGTAGA-3′; reverse: 5′-AGTGCAGGGTCCGAGGTATTC-3′.
Bioinformatic analysis
The target genes of miR-183 were downloaded from the Targetscan database (http://www.targetscan.org/vert_50/) and all verified genes were selected. To explore the functional annotation enrichment of exosomal miR-183 target genes, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using the String database (http://string-db.org/). All target genes of miR-183 were then used to construct a protein-protein interaction network via the String database, with the aim of obtaining a comprehensive description of cellular mechanisms and functions.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM Corp., Armonk, NY, USA). miRNA microarray data were analyzed using a paired sample t-test. The comparison of multiple groups was determined using one-way analysis of variance followed by an S-N-K post-hoc test. P<0.05 was considered to indicate a statistically significant result.
Results
Isolation of plasma-derived exosomes
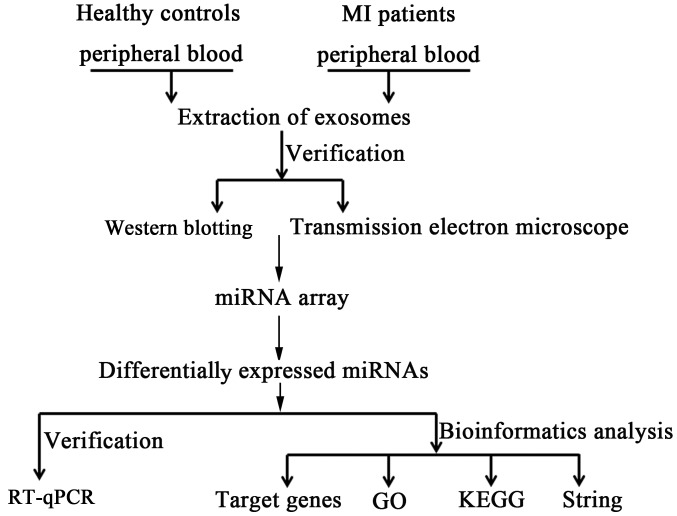
The experimental workflow of the current study is presented in Fig. 1. The presence of extracellular vesicles in patient plasma samples was confirmed using electron microscopy and western blotting. The electron micrographs of harvested extracellular vesicles revealed circular membrane-bound structures with diameters varying from 50 to 100 nm (Fig. 2A). According to these results, it was confirmed that exosomes were successfully isolated from human plasma.
Figure 1.
Diagram of the experiment workflow. MI, myocardial infarction; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
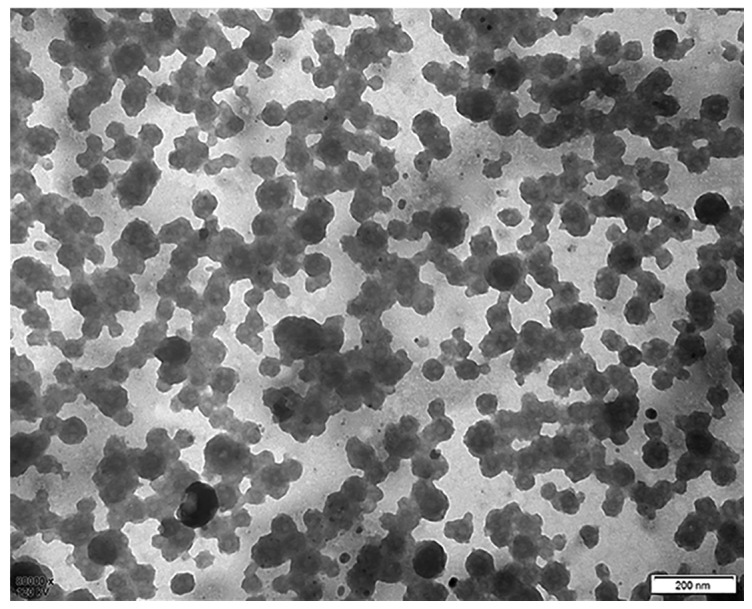
Exsomes identified by electron microscopy. Exosomal morphology was observed using electron microscope.
Expression level of exosomal miR-183 is upregulated in patients with MI
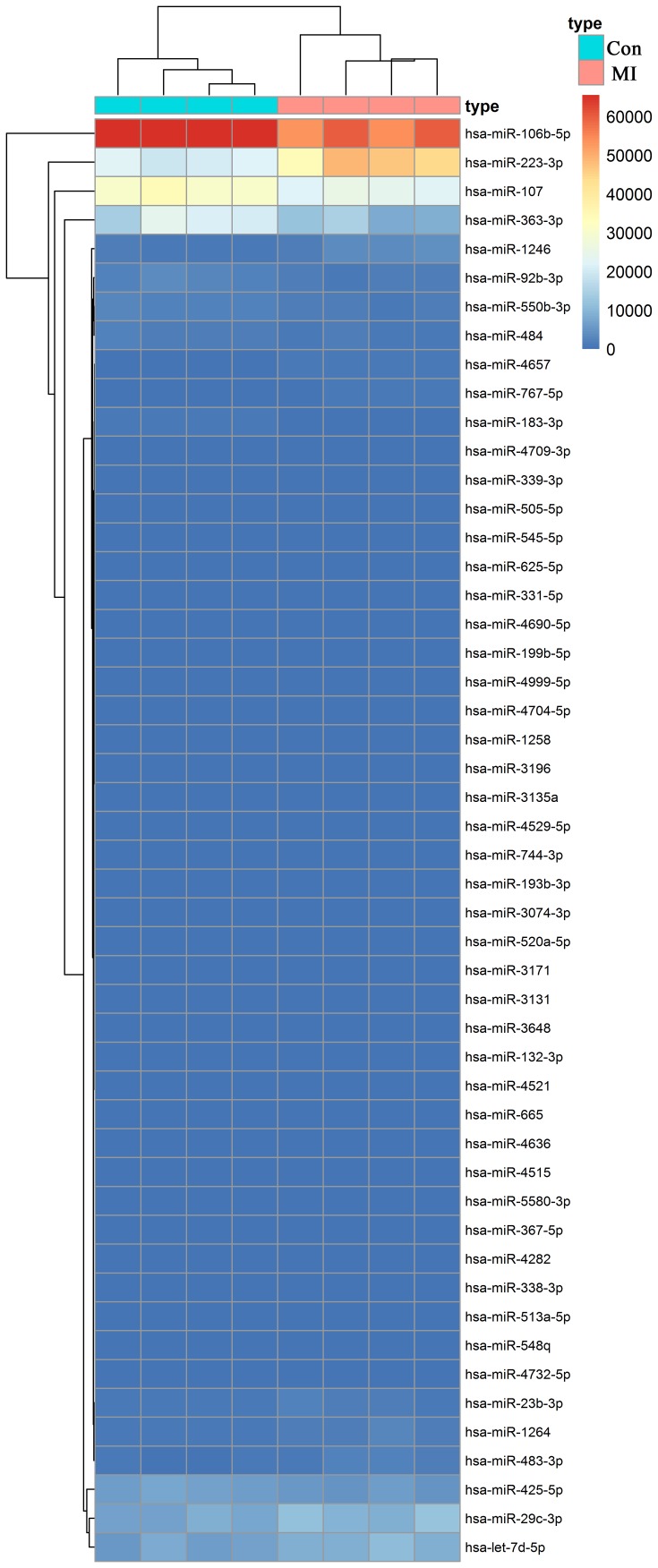
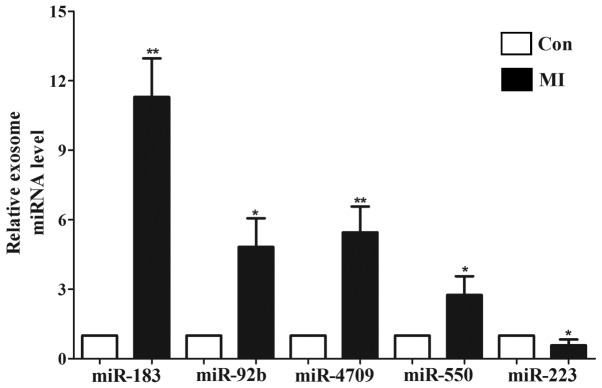
To identify the differentially expressed exosomal miRNAs associated with the occurrence and development of myocardial ischemic injury, a miRNA microarray was used in the present study. The relative expression level of exosomal miRNAs in patients with MI (n=4) and healthy controls (n=4) were compared using a miRNA microarray, from which 85 differentially expressed miRNAs were identified (P<0.05; 1<log2FC<-1; data not shown). The heatmap of the top 50 differentially expressed miRNAs is presented in Fig. 3. The statistical results of 42 significantly different miRNAs (P<0.01) are presented in Table I. The top 10 most markedly dysregulated miRNAs were verified using RT-qPCR in plasma samples from patients with MI (n=14) and healthy individuals (n=14). Five miRNAs were consistent with those of the microarray results: miR-183, miR-92b, miR-4709, miR-550 and miR-223 (Fig. 4). Furthermore, the change in miR-183 expression was the most significant (P<0.01).
Figure 3.
Heatmap of differentially expressed exosomal miRNAs in patients with myocardial infarction and healthy individuals. Con, healthy individuals, MI, myocardial infarction; miR, microRNA.
Table I.
Differentially expressed exosomal miRNAs in patients with myocardial infarction patients and healthy individuals.
| No. | miRNA | Fold-change | Average expression | t-value | P-value |
|---|---|---|---|---|---|
| 1 | hsa-miR-183-3p | 30.4197969 | 517.4346016 | 8.137940658 | 0.000104492 |
| 2 | hsa-miR-4657 | −24.922 | 548.13275 | −7.9080916 | 0.000124557 |
| 3 | hsa-miR-223-3p | −22.225 | 32320.3625 | −7.03659641 | 0.000252489 |
| 4 | hsa-miR-331-5p | 7.78202188 | 98.53995781 | 6.170875344 | 0.000547483 |
| 5 | hsa-miR-3610 | 19.66065609 | −16.82911102 | 6.08685269 | 0.000592724 |
| 6 | hsa-miR-548q | 10.4839219 | 169.6091641 | 5.603577385 | 0.000950949 |
| 7 | hsa-miR-4709-3p | 4.8494688 | 308.1036719 | 5.523099852 | 0.001031681 |
| 8 | hsa-miR-4521 | 3.04811797 | 8.192506641 | 5.340919802 | 0.001244448 |
| 9 | hsa-miR-92b-3p | 14.987188 | 1970.016406 | 5.338298419 | 0.001247849 |
| 10 | hsa-miR-550b-3p | 9.2721875 | 1788.251406 | 5.292485194 | 0.001309002 |
| 11 | hsa-miR-3196 | 4.64892188 | 55.62916875 | 5.164620854 | 0.001498195 |
| 12 | hsa-miR-106b-5p | 8.55 | 61145.74375 | 5.153722569 | 0.001515686 |
| 13 | hsa-miR-4999-5p | 3.437765 | 27.2591675 | 4.881966157 | 0.002035064 |
| 14 | hsa-miR-665 | 2.66090297 | 8.935673828 | 4.686898091 | 0.00253041 |
| 15 | hsa-miR-4636 | 20.69043406 | −14.23388922 | 4.615443949 | 0.002744343 |
| 16 | hsa-miR-505-5p | 9.85890625 | 255.3029219 | 4.580595453 | 0.002855913 |
| 17 | hsa-miR-545-5p | 14.4396688 | 225.7731656 | 4.560897844 | 0.0029212 |
| 18 | hsa-miR-4529-5p | −7.56569 | 16.08826438 | −4.541569 | 0.002986879 |
| 19 | hsa-miR-484 | 7.528 | 1503.836 | 4.504105993 | 0.003118896 |
| 20 | hsa-miR-339-3p | 16.245125 | 293.7521563 | 4.356629784 | 0.003705222 |
| 21 | hsa-miR-4690-5p | 9.08543781 | 87.77904672 | 4.313023159 | 0.003901248 |
| 22 | hsa-miR-4732-5p | 17.3946422 | 156.6910852 | 4.260649424 | 0.004151997 |
| 23 | hsa-miR-199b-5p | −4.177 | 52.0643375 | −4.08801447 | 0.005113256 |
| 24 | hsa-miR-4704-5p | 2.50220313 | 27.57288906 | 4.0354906 | 0.005452571 |
| 25 | hsa-miR-23b-3p | −7.8964844 | 1120.02457 | −4.0042717 | 0.005665949 |
| 26 | hsa-miR-1258 | 28.15745063 | 34.60022938 | 3.971930513 | 0.005896726 |
| 27 | hsa-miR-625-5p | 12.6166406 | 250.8808047 | 3.941044287 | 0.006126798 |
| 28 | hsa-miR-4515 | 17.91870313 | −30.97231094 | 3.917868717 | 0.006305908 |
| 29 | hsa-miR-1264 | −9.83 | 1324.7825 | −3.89505726 | 0.006487833 |
| 30 | hsa-miR-193b-3p | −3.86354734 | 6.368044609 | −3.83238497 | 0.007017982 |
| 31 | hsa-miR-5580-3p | 13.43604625 | −23.61720031 | 3.8175008 | 0.007150752 |
| 32 | hsa-miR-367-5p | −14.87524359 | −15.89417508 | −3.8087925 | 0.007229706 |
| 33 | hsa-miR-363-3p | 9.45 | 15382.3 | 3.778905829 | 0.007508021 |
| 34 | hsa-miR-1246 | −19.130625 | 2229.092188 | −3.74986284 | 0.007789749 |
| 35 | hsa-miR-3131 | 16.80226016 | −0.828861328 | 3.733061229 | 0.007957996 |
| 36 | hsa-miR-3135a | 3.89429531 | 46.65076797 | 3.690694624 | 0.008400146 |
| 37 | hsa-miR-29c-3p | −29.9475 | 8943.44375 | −3.61090831 | 0.00930743 |
| 38 | hsa-let-7d-5p | −26.55 | 7891.1825 | −3.60204751 | 0.009414606 |
| 39 | hsa-miR-3074-3p | −23.74049453 | 13.19788555 | −3.59284785 | 0.009527305 |
| 40 | hsa-miR-3648 | 18.56568 | −3.532107031 | 3.586636253 | 0.009604231 |
| 41 | hsa-miR-520a-5p | −9.91408813 | 20.38453563 | −3.57706628 | 0.009724074 |
| 42 | hsa-miR-338-3p | −11.5395641 | 104.142593 | −3.57291933 | 0.00977651 |
miR or miRNA, microRNA
Figure 4.
Significantly differentially expressed exosomal miRNAs were assessed using reverse transcription-quantitative polymerase chain reaction. *P<0.05, **P<0.01 vs. controls. Con, healthy individuals; MI, myocardial infarction; miR, microRNA.
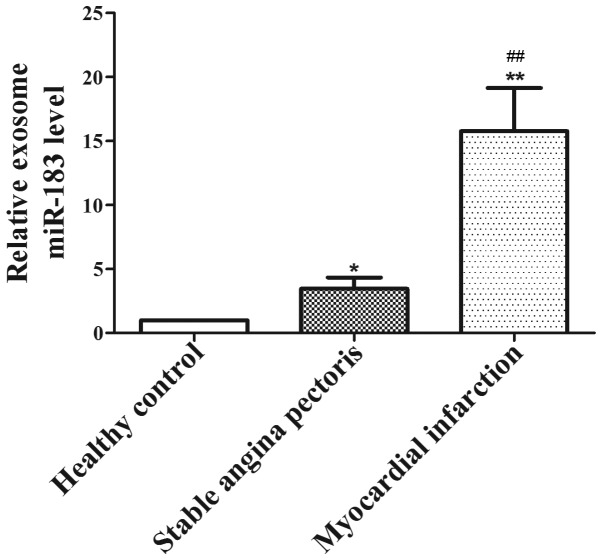
Upregulated miR-183 is associated with the degree of myocardial ischemic injury
To elucidate the role of miR-183 in myocardial injury, the expression of miR-183 in different degrees of myocardial injury was analyzed. The results revealed that exosomal miR-183 levels were significantly lower in the plasma of the healthy individuals (P<0.05) compared with the stable angina and MI groups, and increased accordingly with an increase in severity of myocardial ischemic injury. Furthermore, the expression of miR-183 in patients with MI (n=14) was significantly higher than the levels in patients with stable angina pectoris (n=10; P<0.01; Fig. 5). These results indicate that upregulated exosomal miR-183 may be a novel predictive marker for patients with myocardial ischemic injury.
Figure 5.
Expression of exosomal miR-183 in different degrees of myocardial ischemic injury. *P<0.05, **P<0.01 vs. healthy controls; ##P<0.01 vs. stable angina pectoris. miR, microRNA.
miR-183 is involved in the regulation of protein kinase activity
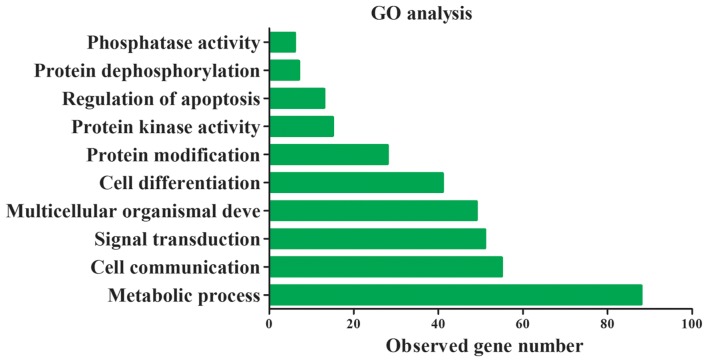
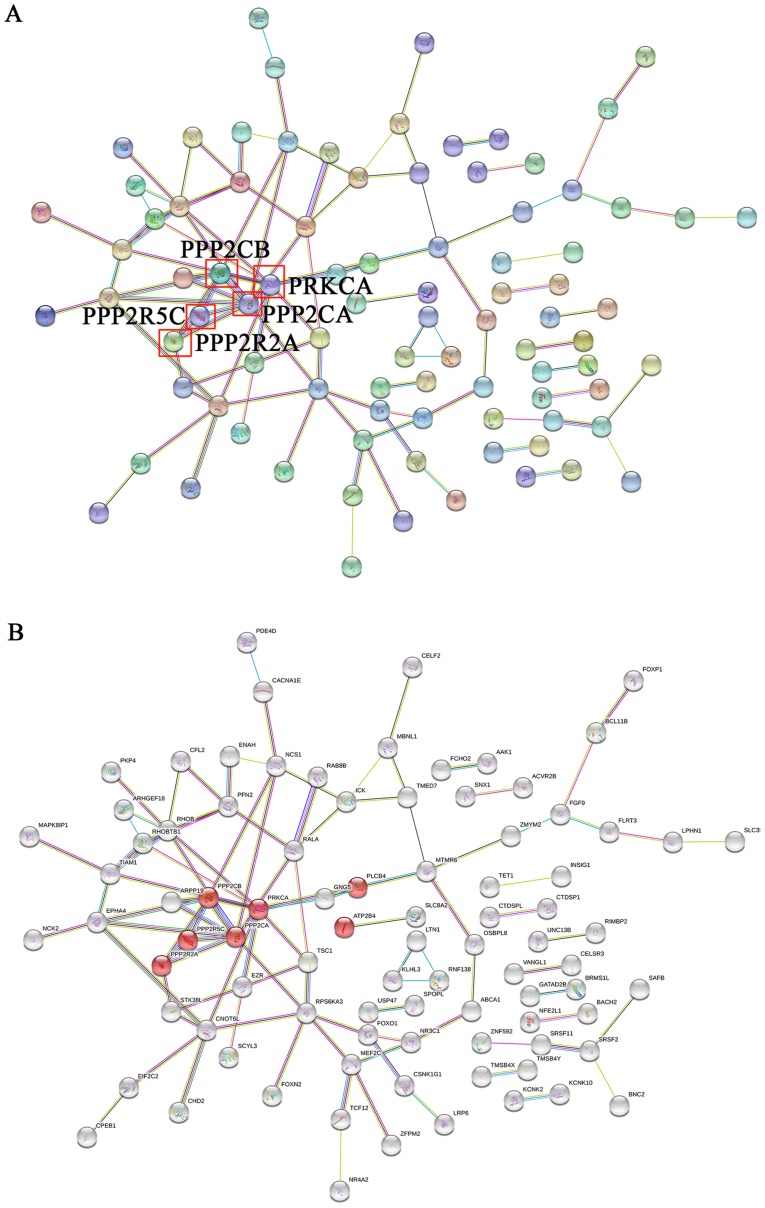
miRNAs are closely associated with heart disease occurrence and progression through the activation of certain signaling pathways (14). To further understand the underlying molecular mechanism of miR-183 function, target genes of miR-193 in GO and KEGG pathways were assessed. The top 30 target genes of miR-183 are presented in Table II. miR-183 target genes were primarily involved in 10 significant functions, including cell communication, signal transduction and the regulation of kinase and phosphatase activity (Fig. 6). The results of KEGG indicate that these target genes are also primarily involved in cardiomyocyte and dopaminergic synapse adrenergic signaling (Table III). A protein-protein interaction network was constructed using the String database and the results demonstrated that the core genes of the network were PPP2CB, PPP2R5C, PPP2R2A, PPP2CA and PRKCA (Fig. 7A). Furthermore, these key genes code for proteins of the kinase family and are involved in adrenergic signaling in cardiomyocytes (Fig. 7B). These results indicate that exosomal miR-183 is involved in cardiomyocyte adrenergic signaling via the regulation of protein kinase activity.
Table II.
The top 30 target genes of miR-183 with higher scores in the TargetScan database.
| No. | Target gene | Total context score | Context score of the aggregate PCT |
|---|---|---|---|
| 1 | SPRY3 | −0.18 | 0.9 |
| 2 | PFN2 | −0.7 | 0.9 |
| 3 | PSEN2 | −0.52 | 0.85 |
| 4 | PPP2R5C | −0.41 | 0.85 |
| 5 | PPP2CA | −0.49 | 0.85 |
| 6 | PDE4D | −0.3 | 0.85 |
| 7 | OSBPL8 | −0.28 | 0.85 |
| 8 | MAL2 | −0.62 | 0.85 |
| 9 | LRP6 | −0.53 | 0.85 |
| 10 | KCNK10 | −0.46 | 0.85 |
| 11 | IDH2 | −0.4 | 0.85 |
| 12 | FCHO2 | −0.39 | 0.85 |
| 13 | EZR | −0.36 | 0.85 |
| 14 | ENAH | −0.41 | 0.85 |
| 15 | DUSP10 | −0.47 | 0.85 |
| 16 | DGCR2 | −0.22 | 0.85 |
| 17 | CTDSP1 | −0.33 | 0.85 |
| 18 | CHD2 | −0.26 | 0.85 |
| 19 | ARHGAP21 | −0.37 | 0.85 |
| 20 | KLHL28 | −0.31 | 0.84 |
| 21 | C16orf72 | −0.43 | 0.83 |
| 22 | PRKCA | −0.1 | 0.77 |
| 23 | PHF15 | −0.4 | 0.77 |
| 24 | PDCD4 | −0.43 | 0.77 |
| 25 | KIAA0368 | −0.22 | 0.77 |
| 26 | KIAA0182 | −0.23 | 0.77 |
| 27 | DAGLA | −0.19 | 0.77 |
| 28 | CDK5R1 | −0.32 | 0.77 |
| 29 | ARPP-19 | −0.5 | 0.77 |
| 30 | AP3M1 | −0.26 | 0.77 |
miR, microRNA; PCT, preferentially conserved targeting.
Figure 6.
GO analysis of target genes of miR-183. GO, Gene Ontology; miR, microRNA.
Table III.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of miR-183 target genes.
| No. | Pathway description | False discovery rate | Matching proteins in network |
|---|---|---|---|
| 1 | Dopaminergic synapse | 0.0171 | GNG5, PLCB4, PPP2CA, PPP2CB, PPP2R2A, PPP2R5C, PRKCA |
| 2 | Adrenergic signaling in cardiomyocytes | 0.0186 | ATP2B4, PLCB4, PPP2CA, PPP2CB, PPP2R2A, PPP2R5C, PRKCA |
| 3 | Regulation of actin cytoskeleton | 0.0186 | CFL2, ENAH, EZR, FGF9, PFN2, TIAM1, TMSB4X, TMSB4Y |
| 4 | Gastric acid secretion | 0.0186 | EZR, KCNK10, KCNK2, PLCB4, PRKCA |
| 5 | AMPK signaling pathway | 0.0214 | FOXO1, PPP2CA, PPP2CB, PPP2R2A, PPP2R5C, TSC1 |
| 6 | MAPK signaling pathway | 0.0428 | CACNA1E, DUSP10, FGF9, MAP3K4, MAPK8IP1, MEF2C, PRKCA, RPS6KA3 |
| 7 | Long-term depression | 0.0473 | PLCB4, PPP2CA, PPP2CB, PRKCA |
miR, micro-RNA. AMPK, AMP-activated protein kinase; MAPK, mitogen-activated protein kinase.
Figure 7.
PPI analysis of miR-183 target genes. (A) The core genes of the PPI network are presented. (B) Genes of the kinase family and adrenergic signaling in cardiomyocytes are presented. PPI, protein-protein interaction; miR, microRNA.
Discussion
In 2012, the World Health Organization Global Status Report on non-communicable diseases revealed that 7.4 million mortalities were a result of ischemic heart disease (15). Specifically, acute MI has a high morbidity and mortality (16). It is thus necessary to elucidate early diagnostic markers. Circulating miRNAs have become novel biomarkers for the detection of various diseases, including those of the cardiovascular system (3). Previous studies have demonstrated that certain circulating miRNAs are involved in acute MI, including cardiac-specific miRNAs, non-cardiac miRNAs (17) and long non-coding RNA (3). Circulating non-coding RNAs can be used as novel biomarkers in the diagnosis of cardiovascular diseases and can assist in cell communication and signal transduction (3,18). Along with the intensification of exosome research, exosomal miRNAs have become novel diagnostic markers in various diseases (19–21). However, the expression of exosomal miRNAs between patients with MI and healthy individuals is yet to be fully elucidated, and confirming the expression profile of exosomal miRNAs in MI is crucial for performing functional research on miRNAs in MI.
Exosomes are 30–120 nm endocytic membrane-derived vesicles (22) that have been reported to serve crucial roles in the clinical diagnosis of certain conditions (22), including ovarian cancer, lung cancer, cardiovascular disease, colorectal cancer (23), and type 1 diabetes (24). Although these exosomes comprise different RNA species, lipids and proteins (25), miRNA levels are closely associated with disease pathophysiology and are high in plasma exosomes (26). A previous study has demonstrated that damaged cardiac muscle cells release exosomes, which regulate the expression of sarcomeric and ion channel genes involved in cardiac conductance, rhythmicity and automaticity via the release of cardiac-specific miRNAs (27).
The present study demonstrated that 50 exosomal miRNAs were dysregulated in patients with MI. Among the dysregulated exosomal miRNAs, miR-183 expression was significantly upregulated in these patients. Previous studies have revealed that miR-183 is primarily involved in the regulation of cancer cell proliferation, invasion and migration in various types of tumor (28–30). Recently, a study demonstrated that the angiogenic factor AGGF1, effectively treats cardiac hypertrophy and heart failure by regulating miR-183 (31). However, the role of miR-183 in MI diagnosis is still unknown. To the best of our knowledge, the present study was the first to demonstrate that exosomal miR-183 is significantly upregulated in patients with MI.
To assess the association between exosomal miR-183 and myocardial ischemic injury, the expression of exosomal miR-183 was measured in healthy individuals, patients with stable angina pectoris and patients with MI. It was revealed that with the increase in the severity of myocardial injury, the expression of exosomal miR-183 also increased. In particular, the expression of exosomal miR-183 was the highest in patients with MI compared to that in healthy controls and patients with stable angina pectoris. To the best of our knowledge, this is the first study to reveal that exosomal miR-183 levels are positively associated with the degree of myocardial injury. A previous study have demonstrated that miR-1, miR-133a and miR-34a are also associated with myocardial injury severity through the induction of adverse structural remodeling, which impairs cardiac contractile function (32). The results of the integrated bioinformatics analysis of the current study indicated that exosomal miR-183 is primarily involved in cardiomyocyte adrenergic signaling via the regulation of protein kinase activity. To the best of our knowledge, there is no study that assesses the involvement of miR-183 in cardiomyocyte adrenergic signaling via the regulation of protein kinase activity. This should therefore be further assessed in future studies. However, the current study has certain limitations. For example, the effects and mechanisms of exosomal miR-183 on cardiac myocyte function need to be further assessed. In addition, the number of cases was small in the present study and as such, there is a requirement for large-scale clinical validation.
In summary, the present study provides an insight into the dynamic changes in exosomal miR-183 levels observed in the early phase of MI and confirms that exosomal miR-183 expression is positively associated with myocardial ischemic injury. Furthermore, the current study confirmed that exosomal miR-183 is primarily involved in cardiomyocyte adrenergic signaling via the regulation of certain protein kinases. Thus, exosomal miR-183 may be a novel diagnostic biomarker for patients with myocardial ischemic injury.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and YJ was responsible for drafting the manuscript, as well as the analysis and interpretation of data. HC and HY collected the data. WG contributed to design of the current study. All authors read and approved the final manuscript
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (Taiyuan, China). Written informed consent was obtained from each patient or their relatives prior to Collection of plasma samples.
Patient consent for publications
All patients agreed with the publication of this study
Competing interests
The authors declare that they have no competing interests.
References
- 1.Xu W, Yu H, Ma R, Ma L, Liu Q, Shan H, Wu C, Zhang R, Zhou Y, Shan H. Apelin protects against myocardial ischemic injury by inhibiting dynamin-related protein 1. Oncotarget. 2017;8:100034–100044. doi: 10.18632/oncotarget.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giustino G, Dangas GD. Ischemia-reperfusion injury and ischemic post-conditioning in acute myocardial infarction: Lost in translation. Catheter Cardiovasc Interv. 2017;90:1068–1069. doi: 10.1002/ccd.27436. [DOI] [PubMed] [Google Scholar]
- 3.Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, Wang Z, Liu Y, Zhang Y. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem. 2017;44:1497–1508. doi: 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- 4.Willeit P, Welsh P, Evans JDW, Tschiderer L, Boachie C, Jukema JW, Ford I, Trompet S, Stott DJ, Kearney PM, et al. High-sensitivity cardiac troponin concentration and risk of first-ever cardiovascular outcomes in 154,052 participants. J Am Coll Cardiol. 2017;70:558–568. doi: 10.1016/j.jacc.2017.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shemisa K, Bhatt A, Cheeran D, Neeland IJ. Novel biomarkers of subclinical cardiac dysfunction in the general population. Curr Heart Fail Rep. 2017;14:301–310. doi: 10.1007/s11897-017-0342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paiva S, Agbulut O. MiRroring the multiple potentials of MicroRNAs in acute myocardial infarction. Front Cardiovasc Med. 2017;4:73. doi: 10.3389/fcvm.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Li XL, Chen ZR, Chng WJ. Tumor-derived exosomes in colorectal cancer progression and their clinical applications. Oncotarget. 2017;8:100781–100790. doi: 10.18632/oncotarget.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alipoor SD, Mortaz E, Garssen J, Movassaghi M, Mirsaeidi M, Adcock IM. Exosomes and exosomal miRNA in respiratory diseases. Mediators Inflamm. 2016;2016:5628404. doi: 10.1155/2016/5628404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson JL, Adams CD, Antman EM, Bridges CR, Califf RM, Casey DE, Jr, Chavey WE, II, Fesmire FM, Hochman JS, Levin TN, et al. 2012 ACCF/AHA focused update incorporated into the ACCF/AHA 2007 guidelines for the management of patients with unstable angina/non-ST-elevation myocardial infarction: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;61:e179–e347. doi: 10.1016/j.jacc.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 10.O' Flynn N, Timmis A, Henderson R, Rajesh S, Fenu E, Guideline Development Group Management of stable angina: Summary of NICE guidance. BMJ. 2011;343:d4147. doi: 10.1136/bmj.d4147. [DOI] [PubMed] [Google Scholar]
- 11.Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Ruan QF, Xie WG, Chen L, Jiang MJ, Ruan JJ, Ye ZQ. Differential expression of microRNAs in serum of severe burn patients and analysis of the signaling pathway at early stage. Zhonghua Shao Shang Za Zhi. 2017;33:639–643. doi: 10.3760/cma.j.issn.1009-2587.2017.10.012. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Verjans R, Peters T, Beaumont FJ, van Leeuwen R, van Herwaarden T, Verhesen W, Munts C, Bijnen M, Henkens M, Diez J, et al. MicroRNA-221/222 family counteracts myocardial fibrosis in pressure overload-induced heart failure. Hypertension. 2018;71:280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 15.Maarman GJ, Mendham AE, Lamont K, George C. Review of a causal role of fructose-containing sugars in myocardial susceptibility to ischemia/reperfusion injury. Nutr Res. 2017;42:11–19. doi: 10.1016/j.nutres.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Tran HV, Lessard D, Tisminetzky MS, Yarzebski J, Granillo EA, Gore JM, Goldberg R. Trends in length of hospital stay and the impact on prognosis of early discharge after a first uncomplicated acute myocardial infarction. Am J Cardiol. 2018;121:397–402. doi: 10.1016/j.amjcard.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ZH, Sun XY, Li CL, Sun YM, Li J, Wang LF, Li ZQ. miRNA-21 expression in the serum of elderly patients with acute myocardial infarction. Med Sci Monit. 2017;23:5728–5734. doi: 10.12659/MSM.904933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, Chen C, Wang DW. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One. 2014;9:e105734. doi: 10.1371/journal.pone.0105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J, Ding M, Xu K, Yang C, Mao LJ. Exosomes in diagnosis and therapy of prostate cancer. Oncotarget. 2017;8:97693–97700. doi: 10.18632/oncotarget.18532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay P, Moodley K, Moodley J, Mackraj I. Placenta-derived exosomes: Potential biomarkers of preeclampsia. Int J Nanomedicine. 2017;12:8009–8023. doi: 10.2147/IJN.S142732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Yang P. A novel cell-cell communication mechanism in the nervous system: Exosomes. J Neurosci Res. 2018;96:45–52. doi: 10.1002/jnr.24113. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, Xu YM, Huang LF, Wang XZ. Exosomes: Novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086. doi: 10.1155/2015/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8:55632–55645. doi: 10.18632/oncotarget.17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Contreras M, Shah SH, Tamayo A, Robbins PD, Golberg RB, Mendez AJ, Ricordi C. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci Rep. 2017;7:5998. doi: 10.1038/s41598-017-05787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluijter JPG, Davidson SM, Boulanger CM, Buzás EI, de Kleijn DPV, Engel FB, Giricz Z, Hausenloy DJ, Kishore R, Lecour S, et al. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: Position paper from the working group on cellular biology of the heart of the European society of cardiology. Cardiovasc Res. 2018;114:19–34. doi: 10.1093/cvr/cvx211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li DB, Liu JL, Wang W, Li RY, Yu DJ, Lan XY, Li JP. Plasma exosomal miR-422a and miR-125b-2-3p serve as biomarkers for ischemic stroke. Curr Neurovasc Res. 2017;14:330–337. doi: 10.2174/1567202614666171005153434. [DOI] [PubMed] [Google Scholar]
- 27.Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac extracellular vesicles in normal and infarcted heart. Int J Mol Sci. 2016;17(pii):E63. doi: 10.3390/ijms17010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macedo T, Silva-Oliveira RJ, Silva VAO, Vidal DO, Evangelista AF, Marques MMC. Overexpression of mir-183 and mir-494 promotes proliferation and migration in human breast cancer cell lines. Oncol Lett. 2017;14:1054–1060. doi: 10.3892/ol.2017.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lima CR, Gomes CC, Santos MF. Role of microRNAs in endocrine cancer metastasis. Mol Cell Endocrinol. 2017;456:62–75. doi: 10.1016/j.mce.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Buurman R, Skawran B, Lehmann U. hsa-mir-183 is frequently methylated and related to poor survival in human hepatocellular carcinoma. World J Gastroenterol. 2017;23:1568–1575. doi: 10.3748/wjg.v23.i9.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao Y, Lu Q, Hu Z, Yu Y, Chen Q, Wang QK. A non-canonical pathway regulates ER stress signaling and blocks ER stress-induced apoptosis and heart failure. Nat Commun. 2017;8:133. doi: 10.1038/s41467-017-00171-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qipshidze Kelm N, Piell KM, Wang E, Cole MP. MicroRNAs as predictive biomarkers for myocardial injury in aged mice following myocardial infarction. J Cell Physiol. 2018;233:5214–5221. doi: 10.1002/jcp.26283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.