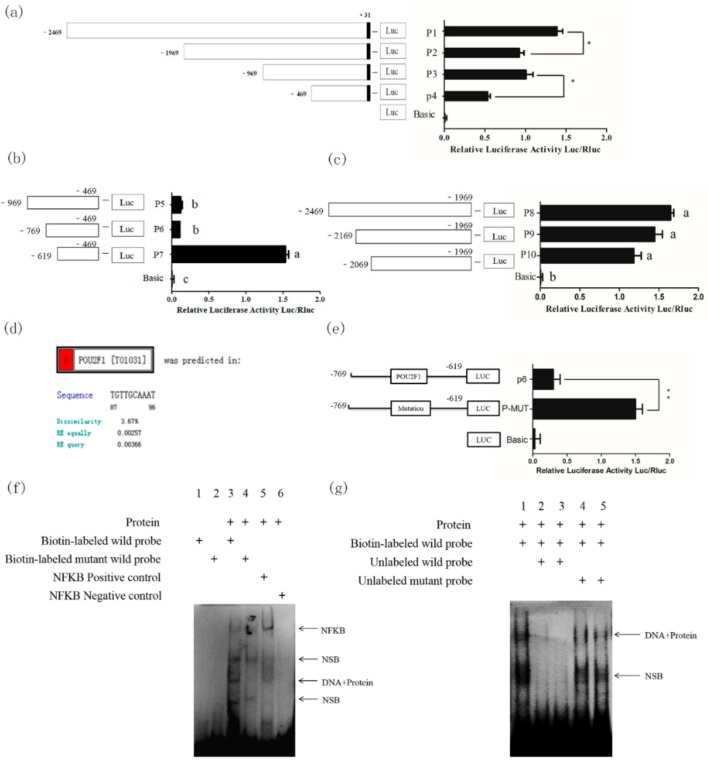
Figure 5.
Regulation of transcriptional factor POU2F1 on Slc7a11 promoter activity. An extremly significant difference was signified with “**” (p < 0.01), and a significant difference was signified with “*” (p < 0.05). (a) Preliminary analysis of the activity of Slc7a11 promoter-deleted vector series. P1~P4 were constructed and the activity of each fragment was detected using dual luciferase. It was presumed that the Slc7a11 promoter contained two active regions, namely −969~−469 bp and −2469~−1969 bp. (b) Activity detection of a series of vectors with deletions in the −969~−469 bp active region of Slc7a11 promoter. P5–P7 were designed and constructed for this region. (c) Activity detection of a series of vectors with deletions in the −2469~−1969 bp active region of Slc7a11 promoter. P8–P10 were designed and constructed for this region. (d) Prediction of the transcriptional binding site in the Slc7a11 promoter region. The −769~−619bp region was the primary target based on the results of (a–c). The results also suggested that transcriptional repressors may be present in this region. The potential transcription factor binding sites were analyzed using the online program PROMO. (e) Site-directed mutagenesis analysis of POU2F1. Based on the predicted position given by PROMO, the POU2F1 binding site was effectively mutated by site-directed mutagenesis and detected by dual-luciferase assay. (f) EMSA suggests that POU2F1 binds to the Slc7a11 core promoter region. The 1st and 2nd lanes were normal and blank mutant probes, respectively, and no bands indicated good probes. The 3rd and 4th lanes were biotin-labeled normal and mutation probes, respectively. The 5th and 6th lanes were NF-KB positive and negative controls, respectively. NSB stands for non-specific binding. (g) Specific binding of POU2F1 to the Slc7a11 core promoter region using competitive EMSA experiments. In the 2nd and 4th lanes, unlabeled probes were added at a 40:1 ratio to the labeled probes, and in 3rd and 5th lanes unlabeled probes were added at an 80:1 ratio to the labeled probes.