Figure 1.
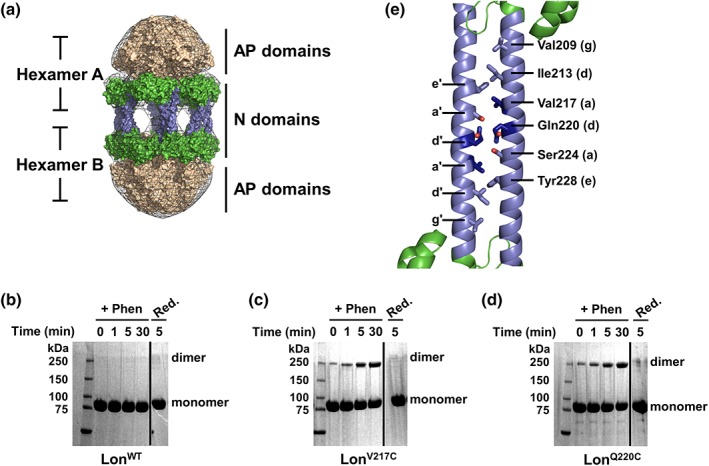
The Lon dodecamer forms via putative N domain coiled‐coil interactions. (a) Low‐resolution electron microscopy 3D reconstruction presents a model for dodecamer formation via the association of Lon N domains through antiparallel coiled coils. Lon N domain dimers are in green with the predicted coiled‐coil region colored blue, the hexameric ATPase and protease domains are colored tan, and the electron density map is shown as a gray mesh. Details for model reconstruction were described previously20. (b–d) Copper phenanthroline catalyzed crosslinking of LonWT and Lon variants analyzed by SDS‐PAGE. Lon proteins (20 μM) were incubated with catalyst (+Phen) for the indicated times, then reduced with 10 mM β‐mercaptoethanol and 5 mM DTT (Red.). LonWT (b) shows no crosslinking in the presence of phenanthroline whereas LonV217C (c) and LonQ220C (d) both show a time‐dependent increase in crosslinking with oxidizing agent. All crosslinking was abolished by addition of reducing agent. Solid line indicates a portion of the gel that was removed for clarity. (e) Bioinformatics analysis was used to create a theoretical model of the Lon N domain antiparallel coiled‐coil interactions (cartoon representation, colored as in (a). The residues chosen for mutagenesis (shown in stick representation) were mapped on the E. coli Lon N domain structure (PDB 3LJC). Two Lon N domain monomers were manually modeled in an antiparallel conformation to mimic the potential interaction surface and residues Val217 and Gln220 are highlighted in dark blue. The lower case letters denote the coiled‐coil heptad position as displayed in Fig. S1.
