Figure 1.
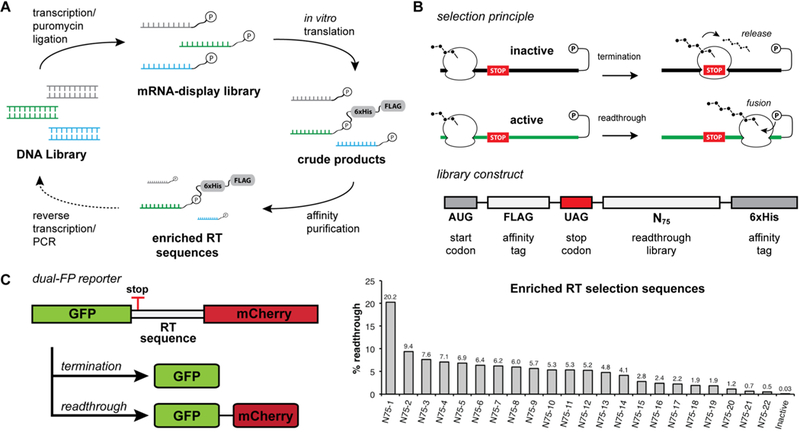
In vitro selection for stop codon readthrough. (A) mRNA display in vitro selection cycle. mRNA is transcribed from the DNA library and then ligated to a puromycin-containing DNA oligonucleotide. The mRNA display library is translated in rabbit reticulocyte lysate, and translation products are affinity purified. Enriched sequences are reverse transcribed and PCR amplified for subsequent rounds of selection. (B) Selection principle and library selection construct. During the mRNA display selection, translation termination at an internal stop codon prevents the formation of the mRNA–peptide fusion and leads to the release of peptides containing affinity tags. Stop codon readthrough allows for translation of the full mRNA template and subsequent fusion of peptide affinity tags to the mRNA template that promotes readthrough. The library selection construct encodes an open reading frame containing an N-terminal FLAG tag, an in-frame UAG stop codon, a library of 75 randomized nucleotides, and a C-terminal hexahistidine tag. (C) Postselection readthrough library sequences were analyzed using a dual-fluorescent protein (dual-FP) reporter assay in Saccharomyces cerevisiae. Readthrough efficiency was determined by comparing the GFP:mCherry fluorescence ratio to a no-stop GFP-mCherry control set to 100%.
