Figure 3.
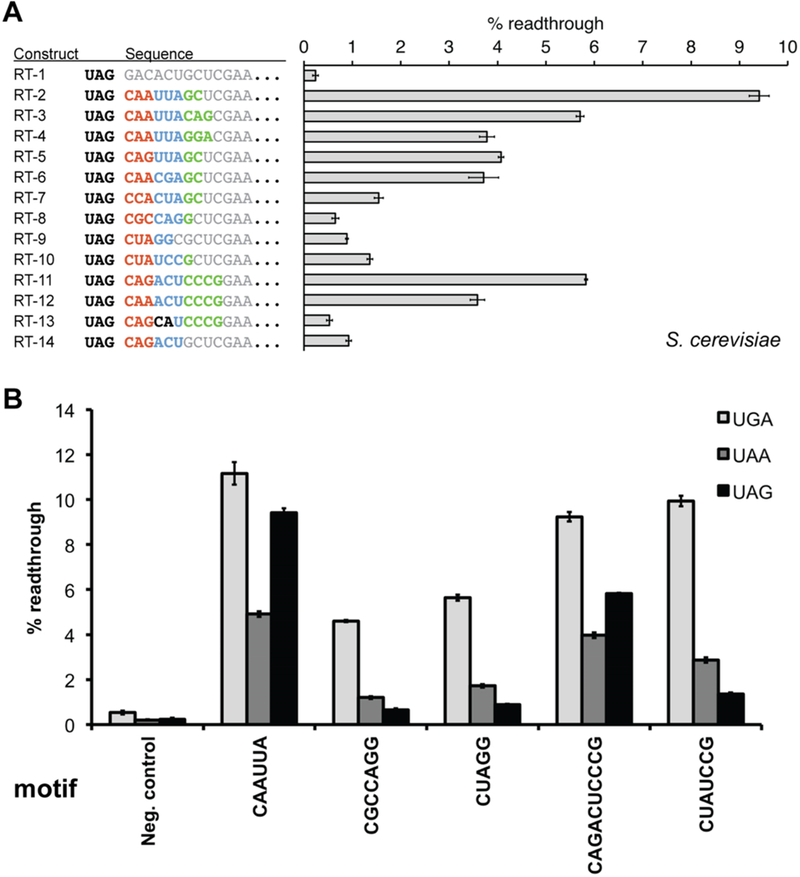
Assays of primary sequence readthrough motifs in yeast. (A) Readthrough sequence motifs recovered from the in vitro selection were evaluated in yeast using the dual-FP reporter assay within a fixed sequence context. RT-1 represents the negative control containing a randomly generated sequence. Sequence elements constituting the RNA motif are shown in color, whereas surrounding fixed sequence context is colored gray. (B) Comparison of primary sequence readthrough motif activities across each of the three stop codons. For all constructs, the readthrough efficiency was determined by comparing the GFP:mCherry fluorescence ratio to a no-stop GFP-mCherry control set to 100%. Neg. control is identical in sequence to RT-1, with only the stop codon substitutions. Error bars represent the standard error of the mean for six or more individual colony replicates.
