Figure 5.
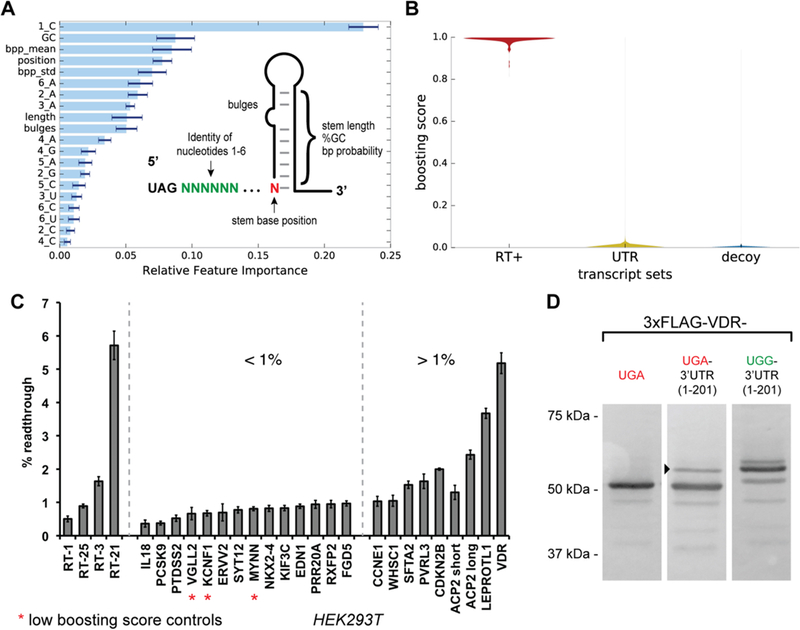
Machine learning prediction of readthrough activity from human transcripts and validation in HEK293T cell assays. (A) The machine learning feature space was constructed from position-specific nucleotide identities and secondary structure attributes. The relative importance of a feature for the additive classifier after training on the readthrough and decoy sequence sets is shown. (B) Violin plot showing the distribution of trained classifier boosting scores for the readthrough (RT+) sequence set, the human 3′-UTR transcript set, and the FLAG decoy sequence set. (C) Selected synthetic readthrough constructs and human 3′-UTRs evaluated for stop codon readthrough in HEK293T cells using the dual-FP reporter assay. Error bars represent the standard error of the mean for three or more independent transfection replicates. (D) Analysis of VDR stop codon readthrough in HEK293T cells. Three constructs were evaluated: full-length VDR containing its native UGA stop codon and no native 3′-UTR sequence, full-length VDR containing its native UGA stop codon and the first 201 nucleotides of its 3′-UTR, and full-length VDR containing a UGA to UGG mutation to its native stop codon and the first 201 nucleotides of its 3′-UTR. The arrowhead indicates the putative readthrough product.
