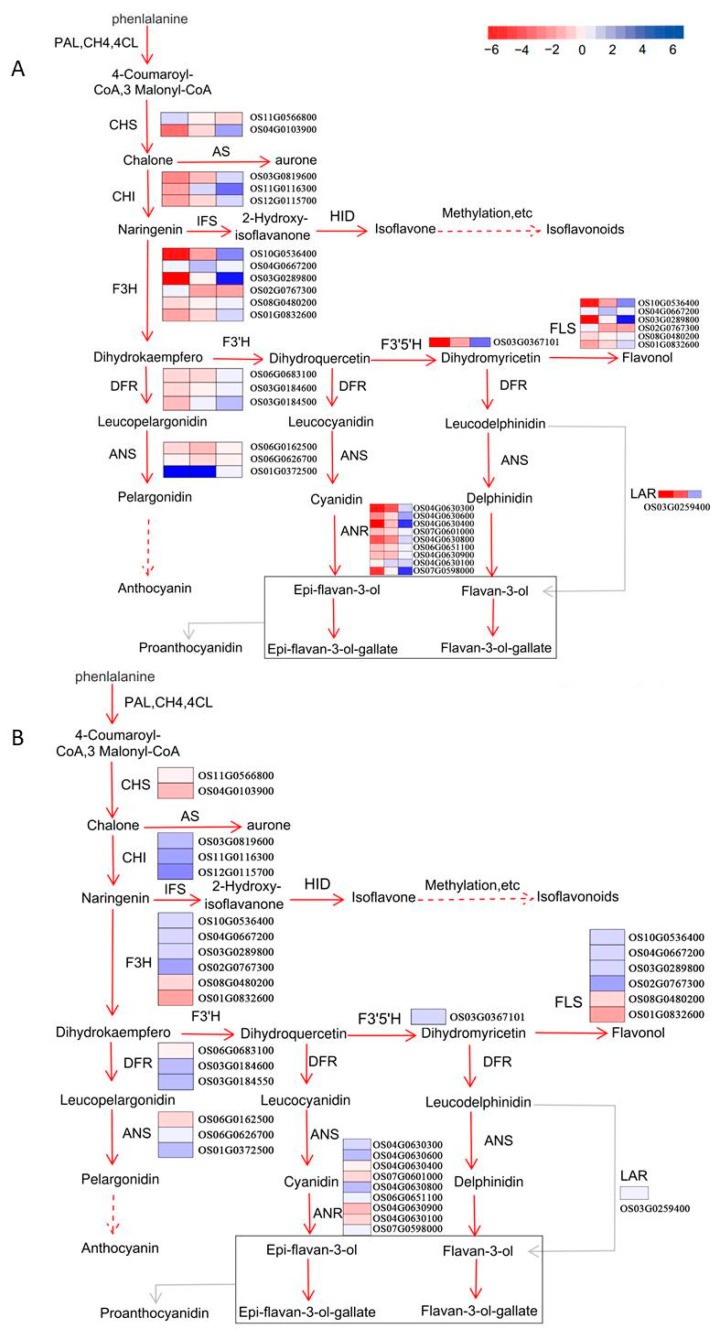
Figure 6.
Pathway map visualizing the changes involved in genes, proteins, and metabolites for the flavonoid biosynthesis pathway among the three rice cultivars. (A) Pathway map for the three cultivars; and (B) pathway map for the pericarp and endosperm of black rice. ANR, anthocyaninidin reductase; ANS, anthocyanin synthase; AS, aurone synthase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; 4CL, 4-coumarate coenzyme A ligase; CHR, chalcone reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3-hydroxylase; F3′,5′H-hydroxylase; FLS, flavonol synthase; HID, 2-hydroxyisoflavanone dehydratase; IFS, 2-hydroxyisoflavanone synthase; LAR, leucoanthocyanidin reductase; PAL, L-phenylalanine ammonia-lyase; and TAL, L-tyrosine ammonia-lyase. Red font represents the key proteins identified by iTRAQ. Blue font represents the key metabolites.