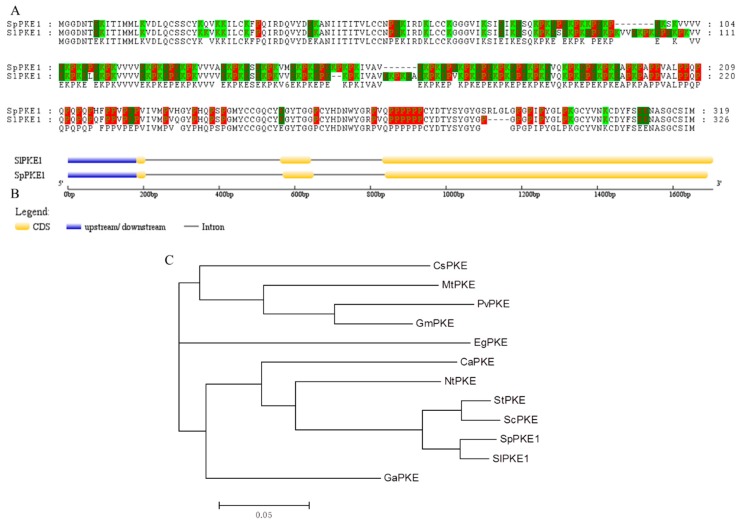
Figure 1.
Comparison of amino acid sequence and phylogenetic analyses of PKE1. (A) Amino acid alignment of S. lycopersicum cv. M82 (SlPKE1) and S. pennellii LA0716 (SpPKE1). (B) Gene structure of the tomato SlPKE1 and SpPKE1 generated from Gene Structure Display Server (GSDS). The yellow block indicates the coding sequence (CDS), the blue block refers to upstream or downstream of the genes, the black line represents the intron. Scale bar indicates the DNA sequence length. (C) Neighbor Joining tree for SlPKE1, SpPKE1 and PKE1 from their highest similarity proteins in the EBI (European Bioinformatics Institute) database. The PKE1 from other species are as follows: CsPKE1 (Citrus sinensis: A0A067GT86), MtPKE1 (Medicago truncatula1: G7JMW9), PvPKE1 (Phaseolus vulgaris: V7CAV2), GmPKE1 (Glycine max: I1MQY4), EgPKE1 (Erythranthe guttata: A0A022QI88), CaPKE1 (Capsicum annuum: Accession no. O81922), NtPKE1 (Nicotiana tabacum: A0A1S4A831), StPKE1 (Solanum tuberosum: M1AFN1), ScPKE1 (Solanum chacoense: A0A0V0I462) and GaPKE1 (Gossypium arboretum: A0A0B0MHK6).