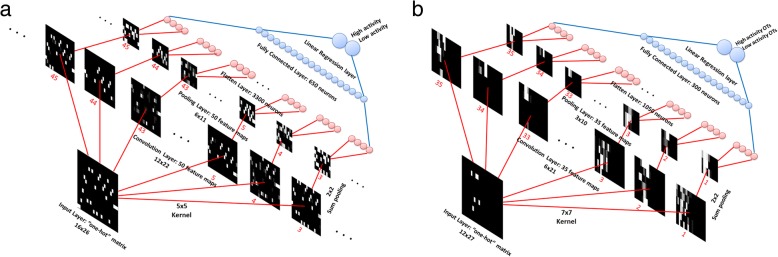
Fig. 1.
Inside the DeepCpf1 architecture. Data flow is from the lower left to upper right. The DNA sequence is translated into a “one-hot” matrix as original input (white indicates 1 and black indicates 0). The convolution and pooling operations are applied to the input and produces the output of each layer as feature maps. The feature maps are visualized as gray scale images by the image function in R. a on-target activity prediction. b off-target specificity prediction