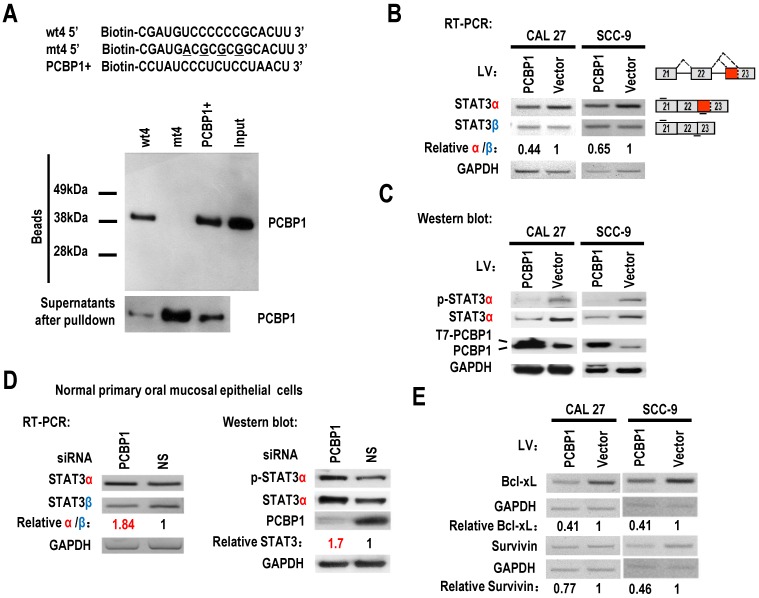
Figure 2.
PCBP1 controls the alternative splicing of STAT3 exon 23. (A) RNA pulldown assay was used to analyze the interaction between PCBP1 and STAT3 RNA. Biotinylated oligo RNAs [including wt4 or mt4 based on minigene mt4, and a positive PCBP1 binding control sequence (PCBP1+)] were incubated with HEK293 total cellular extract. The total proteins binding to RNAs (Beads) were blotted with a mouse anti-PCBP1 antibody. Supernatants after pulldown: total proteins after pulldown in supernatants. (B) CAL 27 or SCC-9 cells were stably transfected with T7 tagged PCBP1 or control lentivirus and the alternative splicing of exon 23 was detected by RT-PCR. Relative α/β represents the ratio of band intensities of α vs β isoform. GAPDH served as a loading control. Diagrams on the right show the structures of STAT3 pre-mRNA and spliced products. Short lines above or below exons stand for primer positions. An exon 22/23 backward junction primer was used to specifically amplify short product, STAT3β. (C) Western blot showed the overexpression of T7 tagged PCBP1 and the expression level of cellular STAT3 and phosphorylated STAT3 (p-STAT3). GAPDH served as a loading control. (D) PCBP1 was knocked down in normal primary oral mucosal epithelial cells. Knockdown efficiency of PCBP1, and the expression of STAT3 and phosphorylated STAT3 were analyzed by western blot. The alternative splicing of exon 23 was detected by RT-PCR. GAPDH served as a loading control. (E) RT-PCR analysis showed that overexpression of PCBP1 downregulates the expression of STAT3 targets (Bcl-xl and Survivin) in both CAL 27 and SCC-9 cells.