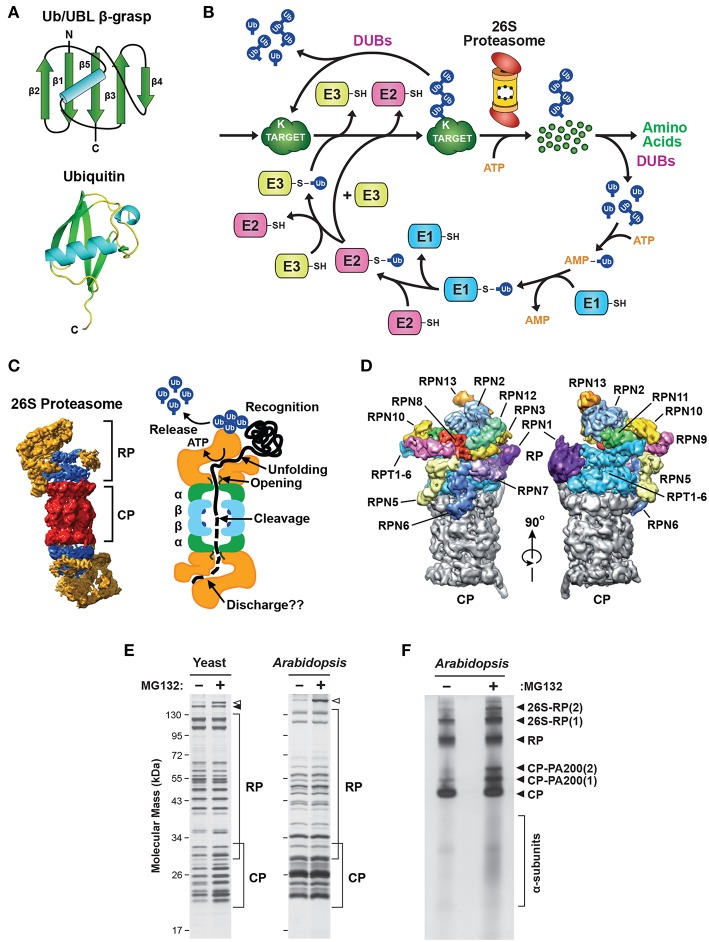
Figure 1.
Description of the Ubiquitin-26S Proteasome System (UPS). (A) The structure of ubiquitin (Ub). Top, schematic of the β-grasp fold of ubiquitin showing the arrangement of the α-helix and β-strand secondary structures. Bottom, a 3-dimensional ribbon diagram of ubiquitin (Protein Data Bank: 1UBQ). C, carboxyl-terminus. (B) Schematic representation of the UPS. The pathway begins with adenosine triphosphate (ATP)-dependent activation of ubiquitin by an E1, followed by transfer of the activated ubiquitin to an E2, and then final attachment of ubiquitin to the target protein with the help of an E3. Typically, the resulting product is a ubiquitin-protein conjugate where the C-terminal glycine carboxyl group of ubiquitin is linked through an isopeptide bond to an accessible ε-amino group of a lysine residue in either the target protein or another ubiquitin molecule. After iterative assembly, the poly-ubiquitylated conjugate can either be disassembled by DUBs, or broken down by the 26S proteasome, in both cases with the concomitant release of the bound ubiquitin moieties intact for re-use. (C) 3-dimensional structure of the yeast 26S holo-proteasome, as determined by cryo-EM (Lasker et al., 2012), with the CP shown in red, the RP base shown in blue, and the RP lid shown in yellow (left), and a cartoon representation of the 26S proteasome, highlighting specific functions of the CP and RP during substrate processing (right). (D) A detailed view of the subunit architecture of the yeast 26S proteasome RP, as determined by cryo-EM (Lander et al., 2012). The CP is shown in gray, the Rpt ring is shown in light blue, and additional Rpn subunits are shown in various colors with their identity indicated. (E) Affinity purification of 26S proteasomes from yeast and Arabidopsis showing the size distribution of core subunits. Yeast cells expressing RPN11-TEV-ProA (left) or Arabidopsis seedlings expressing PAG1-FLAG (right) were treated with or without 50 μM MG132 for 16 h before affinity enrichment of 26S proteasomes based on the Protein A or FLAG tags, respectively. The purified particles were then subjected to SDS-PAGE and stained for protein with silver. The distributions of CP and RP subunits are indicated by the brackets. Open and closed arrowheads locate Blm10 and Ecm29, respectively. (F) Arabidopsis 26S proteasomes affinity-purified as in (E) were separated by native gel electrophoresis and stained for protein with silver. The singly- and doubly-capped 26S complex, and the RP, CP, and CP-PA200 sub-complexes, along with partially assembled CP α-subunit rings, are indicated. Images were adapted with permission from Lander et al. (2012), Lasker et al. (2012), Marshall et al. (2015, 2016), and Marshall and Vierstra (2018a).