Figure 4.
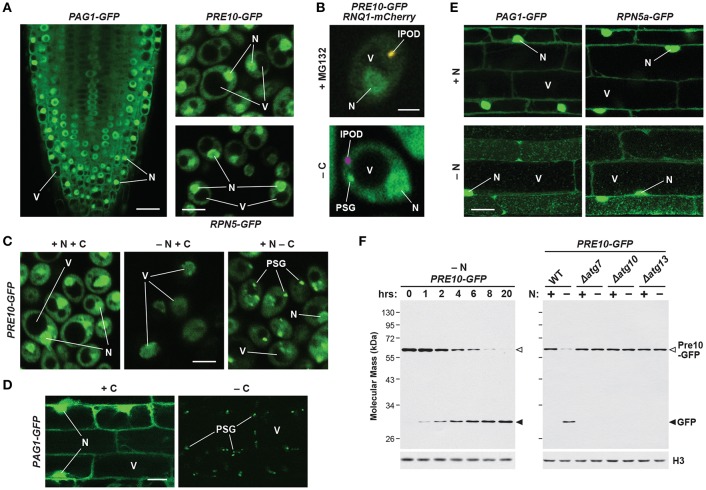
Intracellular Localization of 26S Proteasomes in Arabidopsis and Yeast. The location of proteasomes was tracked by tagging proteasome subunits with GFP, which allows in vivo detection via confocal fluorescence microscopy (A–E), and a quantitative assay for proteaphagy by measuring the release of free GFP from the tagged subunits upon entry into vacuoles (F). (A) 26S proteasomes are found in the cytosol and nucleus of Arabidopsis and yeast cells grown in nutrient-rich conditions. Shown is localization of the PAG1-GFP protein in root tip cells of a 7-day-old Arabidopsis seedling (left), or the Pre10-GFP or Rpn5-GFP proteins in exponential phase yeast cells (top right and bottom right, respectively). Scale bars, 25 μm (left) and 1 μm (right). (B) Yeast 26S proteasomes localize into IPOD-like structures upon inhibition, but to PSGs upon carbon starvation. Cells expressing Pre10-GFP and the IPOD marker Rnq1-mCherry were grown in nutrient-rich medium then switched to either medium containing 80 μM MG132 (top) or medium lacking carbon (bottom) for 8 h and imaged by confocal fluorescence microscopy. Scale bar, 1 μm. (C) Yeast 26S proteasomes are delivered to the vacuole upon nitrogen starvation but sequester into cytoplasmic PSGs upon carbon starvation in yeast. Cells expressing Pre10-GFP were grown in nutrient-rich medium, switched to medium lacking either nitrogen or carbon for 8 h, and then imaged by confocal fluorescence microscopy. Scale bar, 1 μm. (D) 26S proteasomes are sequestered into cytoplasmic PSGs upon fixed carbon starvation in Arabidopsis. 7-day-old Arabidopsis seedlings expressing PAG1-GFP were grown in the light in sucrose-containing medium and then switched to growth in the dark in sucrose-free medium for 16 h. Root cells of the lower elongation zone were imaged by confocal fluorescence microscopy. Scale bar, 10 μm. (E) Arabidopsis 26S proteasomes are sequestered in autophagic bodies inside vacuoles upon nitrogen starvation. Seedlings expressing PAG1-GFP or RPN5a-GFP were grown on nutrient-rich medium and then switched to growth on nitrogen-free medium plus 1 μM concanamycin A for 16 h. Root cells of the lower elongation zone were imaged by confocal fluorescence microscopy. Scale bar, 10 μm. (F) Time course for the autophagy-mediated release of free GFP from Pre10-GFP upon nitrogen starvation in yeast. Wild-type (WT) or autophagy-defective Δatg7, Δatg10, or Δatg13 cells expressing Pre10-GFP were grown in nutrient-rich medium then switched to medium lacking nitrogen for the indicated times (left panel) or 8 h (right panel). Total protein extracts were then assayed for accumulation of free GFP by immunoblot analysis with anti-GFP antibodies. Open and closed arrowheads locate the Pre10-GFP fusion and free GFP, respectively. Immunodetection of histone H3 was used to confirm near-equal protein loading. In panels (A–E); N, nucleus; V, vacuole; IPOD, insoluble protein deposit; PSG, proteasome storage granule. Images were adapted with permission from Marshall et al. (2015, 2016) and Marshall and Vierstra (2018b).