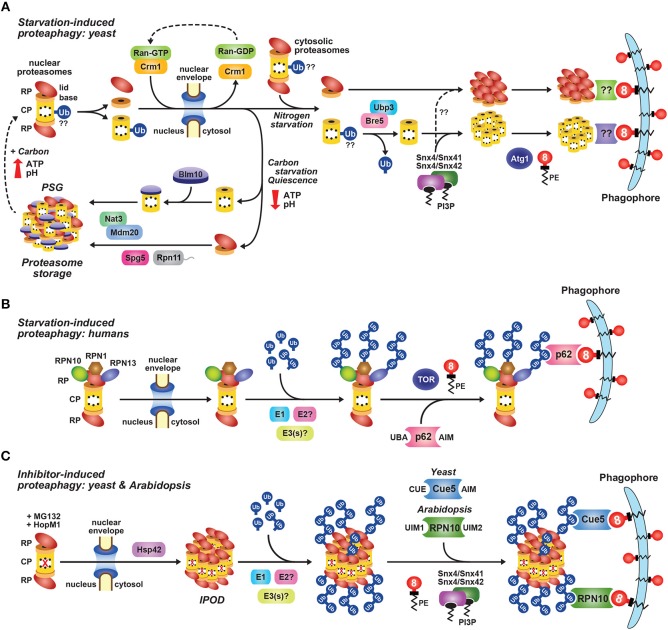
Figure 5.
Pathways for Autophagic Degradation of 26S Proteasomes. (A) A model for starvation-induced proteasome degradation versus storage in yeast. When cells are subjected to nitrogen or carbon starvation, 26S proteasomes dissociate into the CP and RP sub-complexes and are exported from the nucleus via the exportin Crm1. Upon nitrogen starvation, the CP and RP coalesce into cytoplasmic foci in a Snx4/41/42-dependent manner. They are then encapsulated by the expanding phagophore and delivered to the vacuole for degradation, a process requiring the Atg1 kinase complex and the Atg8 lipidation machinery. Deubiquitylation of one or more CP subunits by Ubp3/Bre5 may also be required. Whether specific Atg8-binding autophagy receptors are involved remains unknown. In contrast, carbon starvation, which results in cytoplasmic acidification and reduced ATP levels, triggers re-localization of the CP and RP into cytoplasmic proteasome storage granules (PSGs). This accretion requires numerous factors, including Blm10 for the CP, Spg5 and the C-terminus of Rpn11 for the RP, and the NatB N-terminal acetylation complex (consisting of Nat3 and Mdm20) for both. PSGs act to store proteasome sub-complexes and protect them from autophagic degradation. Preventing sequestration of proteasomes into PSGs leads to their Atg1- and Atg8-dependent turnover. (B) A model for starvation-induced proteasome degradation in humans. When HeLa cells are subjected to amino acid starvation, the proteasome subunits RPN1, RPN10, and RPN13 become poly-ubiquitylated by one or more E3 ligases, facilitating their recognition by the autophagy receptor p62/SQSTM1. By simultaneous interaction with lipidated ATG8/LC3, p62 delivers inactive proteasomes to the expanding phagophore for eventual turnover by autophagy, a process requiring the TOR kinase and the ATG8/LC3 lipidation machinery. (C) A model for inhibitor-induced proteaphagy in Arabidopsis and yeast. Proteasomes subjected to chemical or genetic inhibition, including by the pathogen effector HopM1, are exported from the nucleus and aggregate in an Hsp42-dependent manner into insoluble protein deposit (IPOD)-like structures that are distinct from PSGs. The aggregated proteasomes are then ubiquitylated by one or more E3 ligases, facilitating their recognition by the selective proteaphagy receptors Cue5 in yeast or RPN10 in Arabidopsis. By simultaneous interactions with lipidated ATG8, these receptors deliver inactive proteasomes to enveloping autophagic vesicles for final turnover in the vacuole. PE, phosphatidylethanolamine; PI3P, phosphatidylinositol-3-phosphate.