Table 1.
Examples of MicroED structures
| Sample | Year | Resolution | Ref. | Commentary | |
|---|---|---|---|---|---|
| Lysozyme (14.4 kDa) |
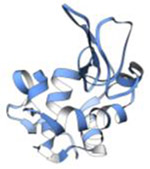 |
2013 | 2.9 Å | 7 | First structure determined by electron diffraction of 3D crystals. PDB ID: 3J4G |
| Lysozyme (14.4 kDa) |
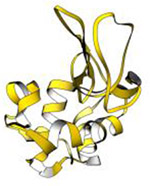 |
2014 | 2.5 Å | 8 | First structure determined by continuous rotation data collection, which currently is the standard MicroED method of data collection. PDB ID: 3J6K |
| Catalase (245 kDa) |
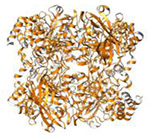 |
2014 | 3.2Å | 33 | These microcrystals resisted structure determination for several decades. PDB ID: 3J7B |
| Sup35 prion protein core (905 Da) |
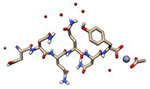 |
2016 | 1.0 Å | 34 | One of the first structures with phases determined by direct methods. PDB ID: 5K2E |
| Trypsin (23.4 kDa) |
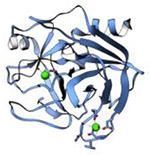 |
2017 | 1.7 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7R |
| Xylanase (21.1 kDa) |
 |
2017 | 2.3 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7P |
| Thaumatin (22.2 kDa) |
 |
2017 | 2.5 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7Q |
| TGF-b/TbRII complex (22.9 kDa) |
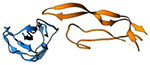 |
2017 | 2.9 Å | 24 | First structure of a protein complex determined by MicroED. PDB ID: 5TY4 |
| Au146(p-MBA)57 nanoparticle (37.5 kDa) |
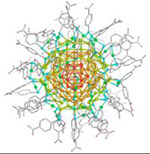 |
2017 | 0.85 Å | 36 | Structure of a gold nanoparticle determined by MicroED |
| Proteinase K (28.9 kDa) |
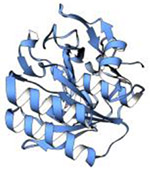 |
2018 | 1.7 Å | 83 | Ultra-low dose MicroED structure. PDB ID: 6CL7 |
| Bank vole prion protein segment (1.1 kDa) |
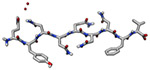 |
2018 | 0.75 Å | 39 | One of the highest resolution MicroED structures to date. PDB ID: 6AXZ |
| Carbamazepine (236 Da) |
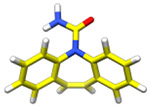 |
2018 | 0.85 Å | 39, 67, 91 | Structure of the drug, carbamazepine determined by MicroED |
