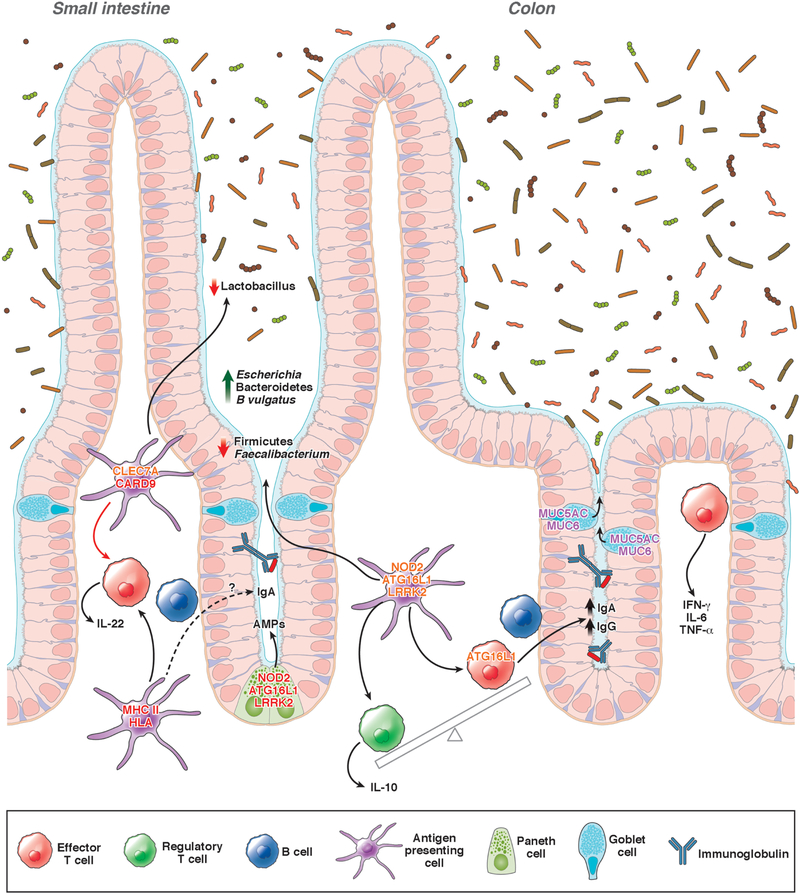
Figure 1. Genetic factors that affect the intestinal microbiome.
Variants genes that affect risk for IBD have been associated with alterations in the composition of the microbiome. Mutations in NOD2, ATG16L1, and LRRK2 reduce secretion of antimicrobial peptide (AMP) by Paneth cells. Variants in CLEC7A and CARD9 have been associated with decreased abundance of Lactobacillus, possibly due to altered activities of dendritic cells and macrophages. Variants in NOD2 are associated with increased abundance of Escherichia species and Bacteroides vulgatus and reductions in Faecalibacterium species. Impaired ATG16L1 signaling has been associated with increased production of IgG and IgA against commensal microbiota, resulting in a loss of tolerance to intestinal microbes. Polymorphisms in MHC class II or HLA genes affect production of IgA in response to microbes. Defects in mucus production alter the intestinal microbiome and increase susceptibility to colitis. Gene names in red have variants associated with CD and UC; gene names in orange have variants associated with only CD; and gene names in purple have variants associated with only UC.