Abstract
Arabidopsis thaliana’s VirE2-Interacting Protein 1 (VIP1) interacts with Agrobacterium tumefaciens VirE2 protein and regulates stress responses and plant immunity signaling occurring downstream of the Mitogen-Activated Protein Kinase (MPK3) signal transduction pathway. In this study, a full-length cDNA of 972bp encoding HvVIP1 was obtained from barley (Hordeum vulgare L.) leaves. A corresponding 323 amino acid poly-peptide was shown to carry the conserved bZIP (Basic Leucine Zipper) domain within its 157th and 223rd amino acid residue. 13 non-synonymous SNPs were spotted within the HvVIP1 bZIP domain sequence when compared with AtVIP1. Moreover, minor differences in the bZIP domain locations and lengths were noted when comparing Arabidopsis thaliana and Hordeum vulgare VIP1 proteins through the 3D models, structural domain predictions and disorder prediction profiling. The expression of HvVIP1 was stable in barley tissues infected by pathogen (whether Agrobacterium tumefaciens or Fusarium culmorum), but was induced at specific time points. We found a strong correlation between the transcript accumulation of HvVIP1 and barley PR- genes HvPR1, HvPR4 and HvPR10, but not with HvPR3 and HvPR5, probably due to low induction of those particular genes. In addition, a gene encoding for a member of the barley MAPK family, HvMPK1, showed significantly higher expression after pathogenic infection of barley cells. Collectively, our results might suggest that early expression of PR genes upon infection in barley cells play a pivotal role in the Agrobacterium-resistance of this plant.
Introduction
Agrobacterium tumefaciens genetically transforms cells by stably integrating a fragment of its extrachromosomal DNA plasmid, the T-DNA, into the plant genome [1]. Molecular and genetic changes occurring within the host organism during the transfection process remain relatively unknown and little is understood about the host factors involved in the process [2–5]. In the model organism Arabidopsis thaliana, the host effector VIP1 interacts with the pathogen’s virulence VirE2 protein (VIP1: VirE2-Interacting Protein 1) and its associated T-complex inside the cell cytosolic space, and guides them into the cell nucleus via its Nuclear Localization Signal motif (NLS), where it then promotes T-DNA integration into the host genome [3, 6–9]. Therefore, VIP1 is thought to be employed by the pathogen to increase the rate of plant genetic transformation. However, VIP1 has been identified as a component of the mitogen-activated protein kinase (MAPK) signal transduction pathway, associated with the regulation of stress responses and plant immunity signalling [10,11]. The current authors attempt to explain and generate theoretical hypotheses and models which can reveal the reasons behind VIP1’s dual function and its controversial behaviours. Our colleagues [4] explained that, during transfection, both the host and Agrobacterium compete in promoting one or other of the two functions of VIP1; “plant susceptibility” to Agrobacterium infection is the determining factor in which role VIP1 actually performs. Thus, they confirmed Karami’s [12] claim that susceptibility to infection remains a genotype-related issue that might vary between tissues and cell types even within the very same plant [13]; more specifically, there may be a correlation with the expression level of certain host proteins [14]. Theoretically, throughout transfection, interaction between VIP1 and bacterial virulence proteins would probably lead the T-DNA to the promoters of particular stress-responsive genes regulated by VIP1; thus, the bacterium’s ability to hijack plant stress response for its own benefits has been highlighted [5,15]. VIP1 (also referred to as bZIP51) protein belongs to a large bZIP family (Basic Leucine Zipper) of transcription factors [16] and has been first characterized in Arabidopsis thaliana [7]. Recently, eight VIP1 homologs have been described in Arabidopsis and tobacco in relation to their interactions with VirE2 [9]. The VIP1 protein sequence has been predicted in wheat (Accession No. AHY03428.1), rice (XP_015618202.1), maize (NP_001141497.1) and barley (BAJ96285.1), but no in-depth study of the protein’s structural nor functional characteristics, in monocotyledonous, which do not host Agrobacterium, has been undertaken.
Pathogenesis-related (PR) genes encoding PR proteins confer pathogen-resistance in host plants. They are usually expressed in plant cells after pathogenic attack or exposure to abiotic stresses, as well as during seed germination, development and senescence [17]. Despite PR proteins being known to possess antifungal activity [18–20], their precise mode of function is still insufficiently understood. In barley, HvPR1, HvPR3, HvPR4, HvPR5 and HvPR10 were found to be induced at different levels following infection with Fusarium culmorum [21], as well as after Agrobacterium infection (as our results have shown). Over the years, studies of the PR1 gene have provided information regarding its roles in plant stress responsiveness. [10] have suggested an MPK3-VIP1-PR1 regulation model in Arabidopsis thaliana cells whereby AtVIP1 transcription factor is considered a direct target of the Agrobacterium-induced mitogen-activated protein kinase (MAPK) MPK3, and such activation of VIP1 by MPK3 will lead eventually to induction of PR1. These findings were subsequently disputed by Pitzschke et al. [11], who claimed that there is no direct interaction between VIP1 and PR1 in reality, suggesting that VIP1’s role in the induction of PR1 expression must occur indirectly. These conflicting results call for resolution through bioinformatic assays or in-vitro protein-DNA binding assays. The barley MAPK family, including 16 MPK proteins, have been characterized previously as key regulators of stress response [22].
The objective of this study was to characterize the VIP1 protein in barley (Hordeum vulgare L., a recalcitrant crop to Agrobacterium-mediated transformation) and to search for structural similarities to and differences from its already characterised Arabidopsis orthologue. Furthermore, we aimed at identifying HvVIP1’s role as a transcriptional regulator of the plant stress response. The expression profile of HvVIP1 as well as other genes implicated in stress (i.e. HvPRs and HvMPK1) were examined in pathogen-treated (i.e. Agrobacterium and Fusarium infected) barley tissues. Furthermore, correlations were examined between HvMPK1 induction and HvVIP1 as well as between HvVIP1 and HvPRs.
Results and discussion
Characterisation of VIP1 in Hordeum vulgare L.
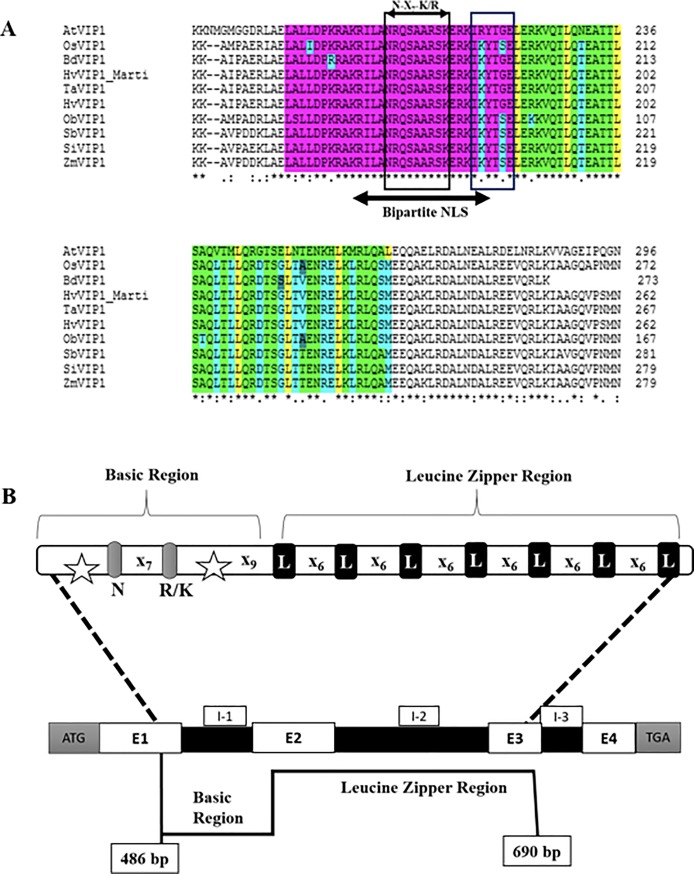
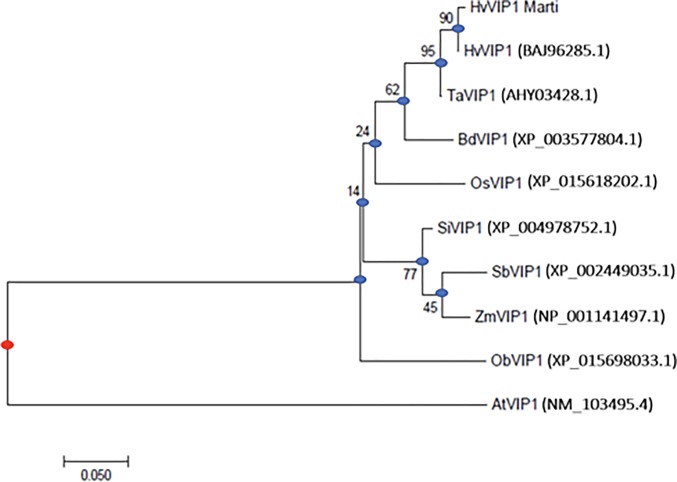
The HvVIP1 full length cDNA in H. vulgare L. cv. Martı is a 972 bp sized fragment encoding a protein of 323 amino acid residues (Accession No: MG000155). Sequence alignment with Hordeum vulgare cv. Haruna nijo predicted clone (DDBJ/EMBL/GenBank accession No. AK365082.1/ [23]) has identified four SNPs within the Martı cv. identified at the 115th, 204th, 872nd, and 896th bp positions, respectively. The HvVIP1 protein sequence is of 323 amino acid residues containing a bZIP domain located near the protein’s C-Terminal Domain (CTD). Based on VIP1-like protein homologies and comparative analysis, the Basic Region (BR) of the bZIP domain was predicted to lie between the 157th and the 187th amino acid. It contains a Nuclear Localization Signal (NLS) sequence and an invariant DNA binding motif. The NLS consensus consists of two short stretches (bipartite) of basic amino acids, designated 165KRILANRQSAARSKERKIK183, which belongs most probably to the class six bipartite NLS model KR-X10-12-K(KR)-X-(K/R) [24]. The DNA binding domain is an invariant N-X7-R/K motif [16] designated within the HvVIP1 protein sequence as 170NRQSAARSK178, marked by the bZIP Group I characteristic Lysine (K) residue at the 178th position replacing the usual Arginine (R) amino acid identified in all other bZIP protein family members. The hinge region located at the end of the Basic Region usually encompasses 6 amino acid residues. It is known for its additive effect upon protein DNA-binding specificity. The Leucine-Zipper (LZ) region is a conserved leucine heptad module located near the protein’s C-terminal domain. It is flanked by the 188th and 223rd residue, marking the end of the bZIP domain, when compared to AtVIP1 [11,16] (Fig 1A). Gene prediction analysis indicated that the HvVIP1 gene is located on the 4th chromosome and has 3620 bp (excluding UTRs). The analyses show that it contains 4 exons of 495 bp, 132 bp, 87 bp and 258 bp size respectively, intercalated by 3 introns of 215 bp, 1691 bp, and 148 bp-sized fragments, respectively. The bZIP domain stretches along Exon 1, 2, and 3, as indicated in the HvVIP1 gene model (Fig 1B). A phylogenetic tree was constructed using the complete amino acid sequences of VIP1-like bZIP proteins from a variety of monocotyledonous and dicotyledonous species. As a result, the VIP1 proteins from cereals were clustered into groups that each of them showed variation, while this clad was quite distinct from AtVIP1 (Fig 2).
Fig 1.
(A) Alignment of the VIP1 bZIP domains of various homologous plant proteins identified by Blast search. The bZIP domain of VIP1 (DDBJ/EMBL/GenBank accession No. NP_564486.1) was aligned (using the clustalW algorithm- EMBL-EBI/MUSCLE) with similar motifs of its homologues from Oryza sativa (OsVIP1), Brachypodium distachyon (BdVIP1), Hordeum vulgare (Both, predicted clone HvVIP1, accession No. BAJ96285.1 and our predicted protein sequence HvVIP1_Martı), Triticum aestivum (TaVIP1), Oryza brachyantha (ObVIP1), Sorghum bicolor (SbVIP1), Setaria italica (SiVIP1), Zea mays (ZmVIP1). Regions of identity are indicated as follows: (i) The basic region is highlighted in violet, (ii) the bipartite NLS within the basic region is denoted by a horizontal bar in grey below its sequence, (iii)the invariant N-X7-K/R motif location is indicated within the basic region, (iv) the Leucine Zipper region (seven Leucine heptad) is highlighted in yellow/green, (v) SNPs within the bZIP domains are marked individually and were identified according to AtVIP1. (*) indicates fully conserved residues, (:) and (.) indicate less conserved residues. (B) Predicted gene model of HvVIP1. The bZIP domain Basic region stretches over Exons 1 and 2 (E-1 and E-2). Within the basic region: bipartite NLS is marked in stars, and the invariant DNA binding motif (N-X7-R/K). The Leucine Zipper region is located on exons 2 and 3 (E-2 and E-3). I 1–3: Introns; (N): Asparagine; (R): Arginine; (K): Lysine; (L): Leucine; ATG: Start codon; TGA: Stop codon; X: any nucleotide.
Fig 2. Phylogeny of the VIP1-like proteins.
The multiple alignment was generated using MUSCLE software and the phylogenetic tree was built with the MEGA5 software using the JTT matrix-based model (the numbers at the nodes indicate the bootstrap scores). The protein accession numbers are indicated in parentheses. Oryza sativa (OsVIP1), Brachypodium distachyon (BdVIP1), Hordeum vulgare (Both, predicted clone HvVIP1, and our predicted protein sequence HvVIP1_Martı), Triticum aestivum (TaVIP1), Oryza brachyantha (ObVIP1), Sorghum bicolor (SbVIP1), Setaria italica (SiVIP1), Zea mays (ZmVIP1), and Arabidopsis thaliana (AtVIP1). Blue nodes are the putative ancestors; the red node represents the historical ultimate ancestor. The Bar (0.050) is a scale for the amount of genetic change (nucleotide substitutions per site).
The Pfam server predicted a bZIP sequence domain between the 160th and the 221st residues within the HvVIP1 protein. In addition, the sub-sequences of the protein’s ordered regions, predicted using DISOclust, indicated that the ordered region containing the bZIP domain lies between the 146th and the 257th residues. Those results were confirmed with the IntFOLD server, which generated 3D models with high quality estimates, structural domain predictions, and disorder prediction profiles. The IntFOLD server indicated a likely bZIP ordered helical domain located between the 146th and the 257th amino acid residues (S1B Fig); whereas, in AtVIP1, the bZIP motif was registered, according to IntFOLD, between the 187th and the 290th amino acid, as shown in S1A Fig. These results suggest that the HvVIP1 Leucine Zipper region was likely to contain up to nine leucine heptad repeats. The leucines are sometimes substituted by isoleucine, valine, phenylalanine or methionine (e.g. Fig 1A- methionine replacing leucine at the 230th position). The intrinsic disorder prediction profile [25] (S1C Fig) of the full-length H. vulgare VIP1 predicted protein indicates that HvVIP1 is partially disordered at the N and C termini and has an ordered region spanning the146th to the 257th residues.
Overall, amino acid sequences of VIP1 orthologues retrieved from both dicotyledonous (A. thaliana) and monocotyledonous (barley, rice, wheat, maize, etc.) counterparts share certain structural similarities at the level of their ordered regions beholding the reputed bZIP domains [16, 26–28]. In our study, a variety of bioinformatics tools, domain database (i.e. Pfam), and servers (i.e. IntFOLD, DISOclust) were used to elucidate the structural features of HvVIP1 protein and eventually to build a protein model [29,30]. IntFOLD server, an integrated web resource for protein fold recognition [31] provided 3D model quality assessment of the submitted protein, whereas DISOclust server was used for prediction of VIP1 disordered regions (S1 Fig) [25,32]. In the future, it would be interesting to test the structural similarities of protein orthologues in an attempt to reflect on their functionality within different phytosystems.
Expression Profile of HvVIP1 in barley infected by pathogens
Gene expression examined in different barley tissues showed that HvVIP1 is expressed in vascular tissues of plants like its counterpart members in the Group I bZIP proteins. Its expression profile in stress-free barley leaves, roots, immature embryos, coleoptiles and calli (Fig 3A) confirmed earlier findings regarding bZIP transcription factors’ role in vascular developmental and plant growth [16,28].
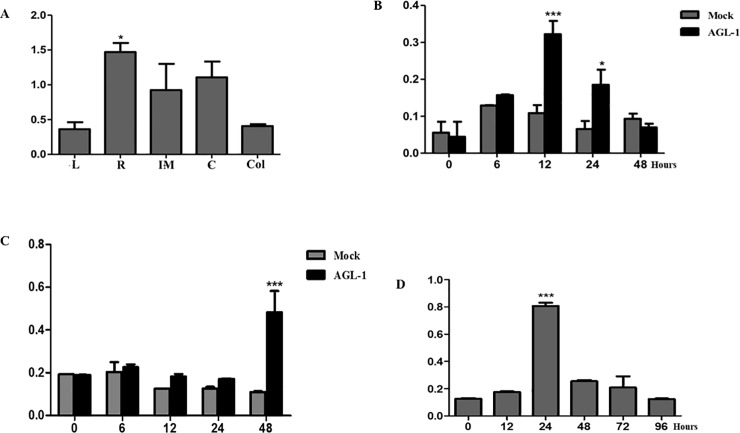
Fig 3. Time-dependent expression of VIP1 in barley cells.
(A) HvVIP1 expression profile in stress-free barley leaves (L), roots (R), immature embryos (IM), calli (C), and coleoptiles (Col). (B) HvVIP1 significantly overexpressed (12th hours) in cv. Golden promise calli after Agrobacterium (AGL-1 strain) infection. (C) HvVIP1 induced significantly at the 48th hour in cv. Martı calli treated with AGL-1 strain. (D) HvVIP1 significantly overexpressed in Fusarium-inoculated barley roots 24 hours after treatment. (*) and (***) designate statistical significance. (*) for P < 0,05; and (***) for P < 0,001.
Infecting barley calli with the Agrobacterium strain AGL-1 led to significant overexpression of HvVIP1 (a 3 fold increase in expression when compared to control/non-inoculated samples) just 12 hours after infection in cv. Golden Promise (susceptible to pathogen infection) (Fig 3B); while HvVIP1 was induced significantly (a 5 fold increase) 48 hours after infection in calli of cv. Martı, a variety known for its pathogen-resistance [21] and tolerance to certain abiotic stresses [33] (Fig 3C). Furthermore, HvVIP1 was induced in barley roots with a significant overexpression (8 fold) observed 24 hours after infection with the Fusarium culmorum F16 strain (Fig 3D).
Based on above-mentioned results, we concluded that VIP1 transcription factor seems to be a stress-induced agent in barley, not only reacting against Agrobacterium, but also responsive to a fungal pathogen.
Previously, VIP1 was described as a transcriptional regulator of stress-responsive genes in wounded and oxidative stress-treated plant cells, as well as a regulator of osmosensory signalling and mechanical stimuli in Arabidopsis thaliana [10,11, 34–36]. In 2008, a study [37] showed that some bZIP family members were upregulated in Arabidopsis by general elicitors of harpin and lipopolysaccharides (LPS), PAMPs that are unique to bacteria; whereas, later on, Lapham and colleagues [38] suggested that AtVIP1 may play a role in fungal (Botrytis cinerea) but not bacterial defense responses in Arabidopsis as it causes the upregulation of two defense related genes (MES1, LYK3) upon fungal attack. Interestingly enough, the recent findings [38] intersect with our results which indicate significant VIP1 induction in barley roots infected (after 24 hours) by another necrotrophic fungus, Fusarium culmorum. Taken together, those results strengthen the role of VIP1 in plant defence response but does not completely clarify it. The contrasting results regarding VIP1 role in defense against bacteria [38] remain to be elucidated by further comparative analyses in dicot and monocot plants.
Expression analysis of HvMPK1 and HvPRs
HvMPK1 is significantly overexpressed in both barley cultivars after bacterial inoculation, particularly 6 hrs after infection (S2A and S2B Fig). Golden Promise calli are more susceptible to infection, with a higher MPK1 induction of up to 500 folds, at the 12th hour. HvMPK1 and HvVIP1 showed a strong correlation, with a correlation coefficient of r = 0.90 (p ≤ 0.05) (Table 1) during Agrobacterium infection. HvMPK1 was also significantly induced under fungal stress even relatively late after root infection (S2C Fig). The induction reaches its peak at 48 hours before it is subsided at 72 hours. No significant correlation was showed between HvMPK1 and HvVIP1 at any point during fungal infection (Table 1).
Table 1. Pearson correlation coefficients between HvVIP1 and other stress responsive genes in barley calli.
| Genes | Agrobacterium infection | Fusarium infection |
|---|---|---|
| HvMPK3 | 0.90* | NS |
| HvPR1 | 0.96** | 0.77* |
| HvPR3 | N/A | N/A |
| HvPR4 | 0.96** | Neg |
| HvPR5 | N/A | N/A |
| HvPR10 | 0.95** | Neg |
(NS) Not significant correlation; (Neg) Negative or downhill correlation; (N/A) Not available data. Significance coefficients
(*) P≤0.05, and
(**) P≤ 0.01
The strong correlation between HvVIP1 and a barley MAPK family member protein, HvMPK1 [22], under bacterial infection in barley cells simulates with earlier findings [11] stating that VIP1 is a component of the MAPK signal transduction pathway, regulating stress-dependent genes in a VRE-dependent manner in Arabidopsis. Indeed, the phosphorylation of VIP1 at Serine 79 residue leads to its nuclear transport in plant cells [10]. Thus, our results reflect on the validity of the proposed “MAPK->VIP1->VRE->gene activation” hypothesis in phytosystems other than the model organism A. thaliana.
On the other side, strongly induced HvMPK1 in Fusarium-treated barley cells was no surprise at all as it reflects on its probable role as a member of stress-responsive MAPKs family member perceiving pathogen elicitors [39]. Even though the MAPK family has not been systematically studied in barley, some of its members are found involved in regulating stress responses citing among others but not limited to barley leaf rust caused by Puccinia hordei infection [22].
Gene expression analysis of HvPR1, HvPR4 and HvPR10 showed that these genes are induced by Agrobacterium infection in a time-dependent manner. Transcript accumulation was higher in Agrobacterium-infected calli than in Fusarium-infected roots (e.g. the induction of HvPR4 in Agrobacterium-treated calli was up to 60 folds increased versus 20 folds only in Fusarium-inoculated roots). In addition, the number of transcripts was significantly higher in tolerant barley (Martı) compared to the sensitive genotype (cv. Golden Promise). The 24th and 48th hours after infection with Agrobacterium were critical for the timing of PR induction. In Martı calli, a strong significant correlation (r = 0.96) was detected between HvVIP1 and HvPR1 (Table 1), while no correlation was found for Golden Promise. We did not observe any detectable expression of HvPR3 and HvPR5 in Agrobacterium-infected barley callus. These genes were also slightly induced in Fusarium-infected roots.
Moreover, HvVIP1 and HvPR1 expression appeared to be harmonized in the Martı root infected with F. culmorum. Both genes are induced 24 hours after infection, as shown in S2D Fig. Indeed, a significant correlation between HvVIP1 and HvPR1 was detected with the Pearson test, revealing a relatively strong positive linear correlation between the two genes tested (r = 0.77).
In a recent study conducted upon VIP2 protein (another VIP protein), authors showed its regulatory role upon NtPR1, a stress-responsive gene, after fungal attack, in tobacco crops [40]. Thus, concluding to the fact that this VIP2-NtPR1 upregulation might be behind Tobacco’s resistance to powdery mildew disease. The TaVIP2-NtPR1 reflect on the HvVIP1-HvPR1 upregulation model but within different pathosystem. The interrelationship between VIP proteins and PR genes should be investigated and their role in conferring plant resistance to pathogen attack (caused by different species of fungi e.g. B. cinerea [41]) must be elucidated.
Testing the VIP1- PRs interrelationship in barley tissues is an original work that lead to the detection of a strong correlation between VIP1 and each of the PRs (PR1, PR4, and PR10) (Table 1). However, this outcome was not the same within barley cultivars (Martı vs. Golden Promise) undergoing the same bacterial infection process. The different expression patterns of plant-defense involved factors among cultivars might be due to genotype-related “Susceptibility” to Agrobacterium [42]. Those results should be re-investigated and supported by further analysis (e.g. search for VRE elements in PR gene promoters, in-vitro protein-DNA binding assays, etc.).
Material and methods
Plant materials and growth conditions
Seeds of barley genotypes cv. Martı and cv. Golden Promise were provided by the Trakya Agricultural Research Institute (Turkey) and USDA (USA) respectively. The Martı cultivar is relatively pathogen- resistant [21], while the Golden Promise cultivar is known for its characteristic susceptibility to pathogens. Three leaf stage plantlets (18 days old) were generated in a hypodroponic system, supplemented with 1X Hoagland solution [43], and grown under controlled conditions (25°C day/night, 40–50% relative humidity, 8h light/16h dark photoperiod with a light intensity of 600 μMol m-2 s-1). Calli were generated from immature embryos sections, grown in the dark under controlled conditions, on MS medium supplemented with 2.5 mg/L Dicamba. Six to eight weeks old calli were selected carefully to proceed with.
Isolation of total RNA and sequencing of HvVIP1 cDNA from barley
Total RNA isolation was performed according to a Trizol-based extraction from barley tissues. Full-length cDNAs of H. vulgare L. were obtained using the “Superscript First-Strand Synthesis System for RT-PCR” commercial kit (Invitrogen, #11904–018). VIP1 gene sequence was mapped through the NCBI Genbank tools, VIP1 transcripts were screened and amplified by PCR analysis using two sets of primers: [(i): F/ 5’ ACAGCGACTTCTCCTTCT 3’; R/ 5’ CCGAACTGTGATCGGTATG 3’; (ii): F/ 5’ TACCGGACTACGCCAAGA 3’; R/ 5’ ACCGAACTGTGATCGGTATG 3’]. Primers were designed in such a way to cover the whole gene coding region and were obtained according to the “Primer quest tools- IDT technologies” software. PCR conditions were 94°C for 3mins; 94°C for 30s; 58°C for 30s; and 72°C for 1min- 34 cycles in total with a 5 mins final elongation step (T100 Thermal Cycle, Bio-Rad, #1861096). Aliquots of individual PCR products (5 μL) were loaded on 1% (w/v) agarose gel and visualized with ethidium bromide staining under UV light. VIP1 cDNA fragments were then cloned following the manufacturer’s instructions (Thermo Scientific InsTAclone PCR Cloning Kit, #K1213). For sequencing, BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems (QVs>20) were used, and GeneMapper IDx v4 software was employed for chromatogram analysis. ORFs and prediction transcription start sites (TSS) were identified using the ORF Finder tool (http://www.ncbi.nlm.nih.gov/gorf.html).
Nucleotide sequence and bioinformatics analysis
Structural similarities between VIP1 orthologues were identified through comparative analyses performed using bioinformatics tools. Full-length sequences of the VIP1-like Proteins from Arabidopsis (NP_564486.1), Barley (BAJ96285.1) and Rice (XP_015618202.1) were initially submitted to the IntFOLD server [29] to obtain 3D models with quality estimates [30], template structures, functional information, structural domain predictions and disorder prediction profiles. Sequences were searched against the Pfam database [44] to obtain sequence domain information. The sub-sequences of the ordered regions, predicted using DISOclust [25], were then taken and submitted to the IntFOLD server to obtain higher quality 3D models [30].
For HvVIP1 gene prediction, the full-length cloned VIP1 nucleotide sequence was searched against the barley genome database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (morexGenes, Accession No: MLOC_71198). Details of the exon and intron gene structure were deduced via sequence alignment assay using the ClustalW2 program (EMBL-EBI/MUSCLE). Phylogenetic analyses were conducted based on the JTT matrix-based model [45] of the Molecular Evolutionary Genetics Analysis software (MEGA7) (http://www.megasoftware.net/mega4/mega.html). Ten amino acid sequences (full-length VIP1 protein sequences) derived from complete coding sequences identified in the genomes of various monocotyledonous (i.e. wheat, maize, rice, etc.) and dicotyledonous species (i.e. Arabidopsis) were compared for their evolutionary relationships and trees were drawn to scale, with branch lengths measured as the number of substitutions per site [46]. MEGA 7 tool using Maximum Likelihood statistical methods were used for this matter.
Plant stress applications
Colonies of A. tumefaciens strain AGL-1 were grown on LB medium (10 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, pH 7.0), supplemented with 20 mg/L rifampicin and 200 mg/L carbenicillin. Colonies reaching a certain log phase (0.8 <OD600 <1) were harvested and the pelleted cells were re-suspended in liquid half-strength MS (Murashige and Skoog) inoculation medium (IM) containing 10 mM MES [2-(N-morpholine)-ethane sulphonic acid], 200 μM acetosyringone, 10g/ L glucose, 1g/L pluronic acid, and 2mM CaCl2, where they grew to a final optical density of OD600 = 1 [47]. Prior to infection, AGL-1 colonies were left to rest in the IM, for 1 hour, in the dark. Eight-week-old fresh embryogenic calli from both cv. Martı (pathogen-resistant) and cv. Golden Promise (pathogen-sensitive) were soaked in the inoculation medium for 30 mins, in the dark, at room temperature [48,49], then left to dry on sterile papers before being transferred into a co-cultivation medium (MS medium supplemented with Dicamba 2.5mg/L, 500μM Acetosyringone, and 800 mg/L freshly prepared L-cystein). Infected calli were laid on co-cultivation medium for 6, 12, 24, and 48 hours, in the dark. Mock-inoculated calli were submerged in an Agrobacterium-free Inoculation Medium (IM Δ AGL-1). They went through the same procedure as the bacterium-inoculated calli. 0 hours represents the point in time before any treatment is performed. Harvested calli from different time points were ground up and preserved at -80°C, prior to any further molecular examination. For the model of Fusarium culmorum infection, the highly pathogenic isolate F. culmorum (F16, also known as F16-5) [50] was used to infect barley roots. The fungal growth and development were performed according to the procedure outlined in [51]. Basically, the strain F16 were cultured on Petri dishes containing Potato Dextrose Agar medium (PDA) and incubated for 7 days under controlled conditions of 25°C for 12/12 h of a day/night cycle and 50% relative humidity, prior to barley root infection. For infection, PDA medium discs (5–6 mm diameter) of the strain F16 were collected and then fixed upon the plantlet’s main root in such a way that the mycelium will be in direct contact with it, at a depth of approximately 1 cm underneath the seed (3-day old plant seedlings). In the control treatments, sterile PDA medium discs were placed on the roots. Samples were collected from both groups (control and infected) at several time points (i.e. 12, 24, 48, 72, and 96 hours). The assay was carried out in a controlled growth chamber (Angelantoni EHC, Massa Martana, PG, Italy) with a light intensity of 600 μmol m-2 s−1 at 25°C, 40% relative humidity and 16/8 h of day/night cycle. The pathogenic inoculation with F16-PDA medium disks of barley roots was already tested and its efficiency was proved according to [21].
Quantitative real time PCR
The total RNA, extracted from barley tissues, was purified and first-strand cDNA fragments of HvVIP1, HvMPK1, and HvPR genes from Hordeum vulgare L. were synthesized, as described earlier. The forward and reverse primers of the genes tested are given in Table 2. Primers were designed through IDT tools, and real-time PCR was performed on a Roche Light Cycler 480. The efficiency of these primers was investigated by applying primer melting curve analysis and gel electrophoresis; both results indicated that each primer pair gave a specific and unique PCR product. Reactions and data analysis were performed as described in [21]. Each reaction consisted of a total volume of 10 μL containing 100 ng of cDNA, 1 U of GoTaq DNA polymerase (Promega, M830), 0.2 mM dNTP, 2.5 mM MgCl2, 0.2 μM of each Forward and Reverse primers specific for each gene (Table 2) and 0.5 μL of 20× Eva Green dye (Biotium, 31000). Each experiment was performed in duplicate and with three biological replicates.
Table 2. Primer sequences for gene expression analysis by qPCR.
| Gene | Accession No | Primers | Annealing Temperature (°C) | Amplicon (bp) |
|---|---|---|---|---|
| HvVIP1 | AK365082 |
5’-GAGAGCTCAAACTCCGGTTG -3’ 5’-GAACTTGACCTGCCGCTATC -3’ |
60 | 114 |
| HvPR1 | X74939 |
5’-CGCCGGGAATGTTGTTGGAC-3’ 5’-ACGAGCGTGCATGAATCCCA-3’ |
60 | 126 |
| HvPR3 | X78672 | 5’-AAGGCCACGTCTCCACCCTA-3’ 5’CAGGTCCGGGTTGCTCACAA-3’ | 58 | 114 |
| HvPR4 | Y10814 |
5’-GCCAAGATCGACACCAACGG-3’ 5’-CAGTCCGAGCTGCTCACTCA-3’ |
60 | 150 |
| HvPR5 | AJ276225 |
5’-ACGACATCTCGGTTATCGACG-3’ 5’-TTATTGCCACTGCAGGCGT-3’ |
60 | 151 |
| HvPR10 | AY220734 |
5’-GCGGCTAGGGTGTTCAAGAC-3’ 5’-CCTGAGCTTCGCCACACAAG-3’ |
60 | 123 |
| HvMPK1 | AK376245 |
5’ -GGTCACCGCCAAGTACAA -3’ 5’ -AAGGCGTTGGCGATCTT -3’ |
59 | 123 |
| HvACT | AK362208 |
5’-CGTGTTGGATTCTGGTGATG -3’ 5’-AGCCAC ATATGCGAGCTTCT -3’ |
60 | 208 |
Conclusion
Our work provides insight into some of the structural and functional features of VIP1 and its orthologues in cereal species that are known to be recalcitrant to Agrobacterium-mediated transformation when compared to the host crop Arabidopsis, however it is clear that further studies are required. We have discovered that HvVIP1 is expressed in barley cells under normal conditions while pathogenic infections induce its transcription temporarily. In addition, there is a transcriptional link between HvVIP1 expression and the induction of HvPR genes. In Arabidopsis, a rapid activation of MPK3 upon stress is followed by VIP1 phosphorylation and nuclear translocation within minutes, as has been shown in earlier studies. Our study confirmed the substantial expression of a MAPK protein, HvMPK1, for the first time in relation to both Agrobacterium and Fusarium-infections. This finding clearly puts MAPK signal transduction as a key factor in the response to stress and subsequent tolerance in barley cells. In the future work, it would be interesting to study the expression patterns of other MPK genes in barley as well as investigate in-depth the regulatory role of HvVIP1 upon stress-responsive genes, such as PRs.
Supporting information
(A) 3D model using the full-length Arabidopsis sequence obtained from the IntFOLD server (McGuffin et al., 2015). The model is coloured according to the ModFOLD6 (Maghrabi et al., 2017) predicted per-residue accuracy using a temperature spectrum from blue-red (blue, high confidence and ordered; red, low confidence & disordered). (B) 3D model using the full-length H. vulgare sequence obtained from the IntFOLD server. (C) Intrinsic disorder prediction profile of the full-length barley sequence, from DISOclust (McGuffin, 2008).
(TIFF)
HvMPK1 expression profile in (A) Agrobacterium-infected cv. Golden promise; (B) in Agrobacterium-infected cv. Martı calli; (C) in Fusarium-inoculated cv. Martı roots. (D) Schematic representation of the correlation pattern (r = 0.77) between HvVIP1 and HvPR1 in Fusarium-inoculated barley roots. (*) and (***) designate statistical significance. (*) for P < 0,05; and (***) for P < 0,001.
(TIFF)
Acknowledgments
The authors thank Dr. Cuneyt Ucarli for his valuable contribution in providing laboratory assistance throughout the study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Scientific Research Projects Unit (BAP) at Istanbul University, Project No 20134. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schell J, Van Montagu M, De Beuckeleer M, De Block M, Depicker A, De Wilde M, et al. Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1979; 204, 1155. [DOI] [PubMed] [Google Scholar]
- 2.Tzfira T., Citovsky V., Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Current Opinion in Biotechnology. 2006; 17(2): 147–154. 10.1016/j.copbio.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 3.Gelvin SB. Agrobacterium-Mediated Plant Transformation: the Biology behind the “Gene-Jockeying” Tool. Microbiology and Molecular Biology Reviews. 2003; 16–37. 10.1128/MMBR.67.1.16-37.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO. 2010; 29: 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacroix B, Citovsky V. The roles of bacterial and host plant factors in Agrobacterium- mediated genetic transformation. International Journal of Developmental Biology. 2013; 57: 467–481. 10.1387/ijdb.130199bl [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birch RG. Plant Transformatıon: Problems and Strategies for Practical Application. Annual Review of Plant Physiology and Plant Molecular Biology. 1997; 48: 297–326. 10.1146/annurev.arplant.48.1.297 [DOI] [PubMed] [Google Scholar]
- 7.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO. 2001; 20: 3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacroix B, Citovsky V. Characterization of VIP1 activity as a transcriptional regulator in vitro and in planta. Scientific Reports. 2013; 3, 2440 10.1038/srep02440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Lacroix B, Guo J, Citovsky V. The Agrobacterium VirE2 effector interacts with multiple members of the Arabidopsis VIP1 protein family. Molecular Plant Pathology. 2018; 19 (5): 1172–1183. 10.1111/mpp.12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signalling. Science. 2007; 318: 453–456. 10.1126/science.1148110 [DOI] [PubMed] [Google Scholar]
- 11.Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen- activated protein kinase 3-induced stress gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009; 106: 18414–18419. 10.1073/pnas.0905599106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karami O, (2008). Factors affecting Agrobacterium-mediated transformation of plants. Transgenic Plant Journal. 2008; 2: 127–137. [Google Scholar]
- 13.Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annual Review of Plant Physiology and Plant Molecular Biology. 2000; 51: 223–56. 10.1146/annurev.arplant.51.1.223 [DOI] [PubMed] [Google Scholar]
- 14.Hwang HH, Gelvin SB. Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell. 2004; 16: 3148–3167. 10.1105/tpc.104.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Kong X, Pan J, Li D. VIP1: Linking Agrobacterium-mediated transformation to plant immunity?. Plant Cell Reports. 2010; 29: 805–812. 10.1007/s00299-010-0870-4 [DOI] [PubMed] [Google Scholar]
- 16.Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends in Plant Science. 2002; 7: 106–111. [DOI] [PubMed] [Google Scholar]
- 17.Van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defence related proteins in infected plants. Annual Review of Phytopathology. 2006; 44: 135–162. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 18.Desmond OJ, Edgar CI, Manners JM, Maclean DJ, Schenk PM, Kazan K. Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiological and Molecular Plant Pathology. 2006; 67: 171–179. [Google Scholar]
- 19.Sudisha J, Sharathchandra RG, Amruthesh KN, Kumar A, Shetty HS. Pathogenesis related proteins in plant defense response. Plant defence: Biological control, progress in biological control. 2012; 379–403. [Google Scholar]
- 20.Agarwal P, Bhatt V, Singh R, Das M, Sopory SK, Chikara J. Pathogenesis-related gene, JcPR-10a from Jatropha curcas exhibit RNase and antifungal activity. Molecular Biotechnology. 2013; 54: 412–425. 10.1007/s12033-012-9579-7 [DOI] [PubMed] [Google Scholar]
- 21.Tufan F, Uçarlı C, Tunali B, Gürel F. Analysis of early events in barley (Hordeum vulgare L.) roots in response to Fusarium culmorum infection. European Journal of Plant Pathology. 2016; 148(2): 343–355. [Google Scholar]
- 22.Krenek P, Niks RE, Vels A, Vyplelova P, Samaj J. Genome-wide analysis of the barley MAPK gene family and its expression patterns in relation to Puccinia hordei infection. Acta Physiologiae Plantarum. 2015; 37: 254. [Google Scholar]
- 23.Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, et al. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiology. 2011; 156(1), 20–28. 10.1104/pp.110.171579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang C, Couñago RM, Williams SJ, Boden M, Kobe B. The distribution of different classes of nuclear localization signals (NLSs) in diverse organisms and the utilization of the minor NLS-binding site inplantnuclear import factor importin-α. Plant Signaling and Behavior. 2013; 8(10), e25976 10.4161/psb.25976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mcguffin LJ. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics. 2008; 24: 1798–1804. 10.1093/bioinformatics/btn326 [DOI] [PubMed] [Google Scholar]
- 26.Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. TIBS20; 1995. [DOI] [PubMed] [Google Scholar]
- 27.Sun MY, Fu XL, Tan QP, Liu L, Chen M, Zhu CY, et al. Analysis of basic leucine zipper genes and their expression during bud dormancy in peach (Prunus persica). Plant Physiology and Biochemistry. 2016; 104: 54–70. 10.1016/j.plaphy.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 28.Ali Z, Sarwat SS, Karim I, Faridi R, Jaskani MJ, Khan AA. Functions of Plant’s bZIP Transcription Factors. Pakistan Journal of Agricultural Sciences. 2016; 53 (2), 303–314. [Google Scholar]
- 29.Mcguffin LJ, Atkins JD, Salehe BR, Shuid AN, Roche DB. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Research. 2015; 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maghrabi AHA, McGuffin LJ. ModFOLD6: an accurate web server for the global and local quality estimation of 3D protein models. Nucleic Acids Research. 2017; 10.1093/nar/gkx332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche DB, Buenavista MT, Tetchner SJ, McGuffin LJ. The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 2011; 39: W171–W176. 10.1093/nar/gkr184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004; 337: 635–645. 10.1016/j.jmb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 33.Gürel F, Öztürk NZ, Yörük E, Uçarlı C, Poyraz N. Comparison of expression patterns of selected drought-responsive genes in barley (Hordeum vulgare L.) under shock dehydration and slow drought treatments. Plant Growth Regulation. 2016; 80 (2): 183–193. [Google Scholar]
- 34.Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, et al. Isolation and characterization of low- sulphur-tolerant mutants of Arabidopsis. Journal of Experimental Botany. 2010; 61: 3407–3422. 10.1093/jxb/erq161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsugama D, Liu S, Takano T. A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiology. 2012; 159: 144–155. 10.1104/pp.112.197020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsugama D, Liu S, Takano T. VIP1 is very important/interesting protein 1 regulating touch responses of Arabidopsis. Plant Signaling and Behavior. 2016; 11(6), e1187358 10.1080/15592324.2016.1187358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livaja M, Zeidler D, Rad UV, D J. Transcriptional responses of Arabidopsis thaliana to the bacteria-derived PAMPs harpin and lipopolysaccharide. Immunobiology. 2008; 213: 161–171. 10.1016/j.imbio.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 38.Lapham R, Lee LY, Tsugama D, Lee S, Mengiste T, Gelvin S. VIP1 and its homologs are not required for Agrobacterium-mediated transformation, but play a role in Botrytis and salt stress responses. Frontiers in Plant Science. 2018; 9: 749 10.3389/fpls.2018.00749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamel LP, Nicole MC, Duplessis S, Ellis BE. Mitogen-activated protein Kinase signalling in plant-interacting fungi: distinct messages from conserved messengers. The Plant Cell. 2012; 24: 1327–51. 10.1105/tpc.112.096156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao P, Wang K, Lin Z, Zhang W, Du L, Zhang Y, Ye X. Cloning and characterization of TaVIP2 gene from Triticum aestivum and functional analysis in Nicotiana tabacum. Scientific Reports. 2016; 6: 37602 10.1038/srep37602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiba A, Nishihara M, Nakatsuka T, Yamamura S. Pathogenesis-related protein 1 homologue is an antifungal protein in Wasabia japonica leaves and confers resistance to Botrytis cinerea in transgenic tobacco. Plant Biotechnol. 2007; 24: 247–253. [Google Scholar]
- 42.Tie W, Zhou F, Wang L, Xie W, Li X, Lin Y. Reasons for lower transformation efficiency in indica rice using Agrobacterium tumefaciens-mediated transformation: lessons from transformation assays and genome-wide expression profiling. Plant Molecular Biology. 2012; 78: 1–18. 10.1007/s11103-011-9842-5 [DOI] [PubMed] [Google Scholar]
- 43.Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. University of California, Agricultural Experiment Station; 1950; 347; 1–32. [Google Scholar]
- 44.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2015; 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones HD, Doherty A, Wu H. Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods. 2005; 1: 1–5. 10.1186/1746-4811-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016; 33:1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joyce P, Kuwahata M, Turner N, Lakshmanan P. Selection system and co-cultivation medium are important determinants of Agrobacterium-mediated transformation of sugarcane. Plant Cell Reports. 2010; 29: 173–183. 10.1007/s00299-009-0810-3 [DOI] [PubMed] [Google Scholar]
- 48.Li DD, Shi W, Deng XX. Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Reports. 2002; 21:153–156. [Google Scholar]
- 49.Hensel G, Kastner C, Oleszczuk S, Riechen J, Kumlehn J. Agrobacterium-mediated gene transfer to cereal crop plants: Current protocols for barley, wheat, triticale, and maize. International Journal of Plant Genomics. 2009; 835608, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tunali B, Nicol J, Erol FY, Altiparmak G. Pathogenicity of Turkish crown and head scab isolates on stem bases om winter wheat under greenhouse conditions. Plant Pathology Journal. 2006; 5(2): 143–149. [Google Scholar]
- 51.Covarelli L, Gardiner D, Beccari G, Nicholson P. Fusarium virulence assay on wheat and barley seedlings. Bio-protocol. 2013; 3(7): e446. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) 3D model using the full-length Arabidopsis sequence obtained from the IntFOLD server (McGuffin et al., 2015). The model is coloured according to the ModFOLD6 (Maghrabi et al., 2017) predicted per-residue accuracy using a temperature spectrum from blue-red (blue, high confidence and ordered; red, low confidence & disordered). (B) 3D model using the full-length H. vulgare sequence obtained from the IntFOLD server. (C) Intrinsic disorder prediction profile of the full-length barley sequence, from DISOclust (McGuffin, 2008).
(TIFF)
HvMPK1 expression profile in (A) Agrobacterium-infected cv. Golden promise; (B) in Agrobacterium-infected cv. Martı calli; (C) in Fusarium-inoculated cv. Martı roots. (D) Schematic representation of the correlation pattern (r = 0.77) between HvVIP1 and HvPR1 in Fusarium-inoculated barley roots. (*) and (***) designate statistical significance. (*) for P < 0,05; and (***) for P < 0,001.
(TIFF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.