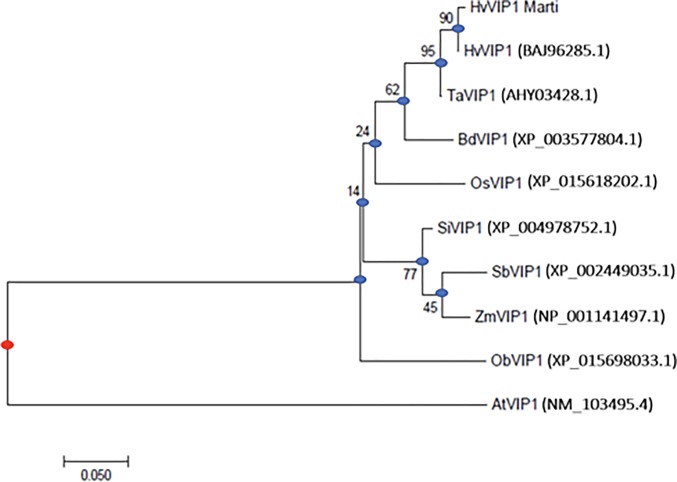
Fig 2. Phylogeny of the VIP1-like proteins.
The multiple alignment was generated using MUSCLE software and the phylogenetic tree was built with the MEGA5 software using the JTT matrix-based model (the numbers at the nodes indicate the bootstrap scores). The protein accession numbers are indicated in parentheses. Oryza sativa (OsVIP1), Brachypodium distachyon (BdVIP1), Hordeum vulgare (Both, predicted clone HvVIP1, and our predicted protein sequence HvVIP1_Martı), Triticum aestivum (TaVIP1), Oryza brachyantha (ObVIP1), Sorghum bicolor (SbVIP1), Setaria italica (SiVIP1), Zea mays (ZmVIP1), and Arabidopsis thaliana (AtVIP1). Blue nodes are the putative ancestors; the red node represents the historical ultimate ancestor. The Bar (0.050) is a scale for the amount of genetic change (nucleotide substitutions per site).