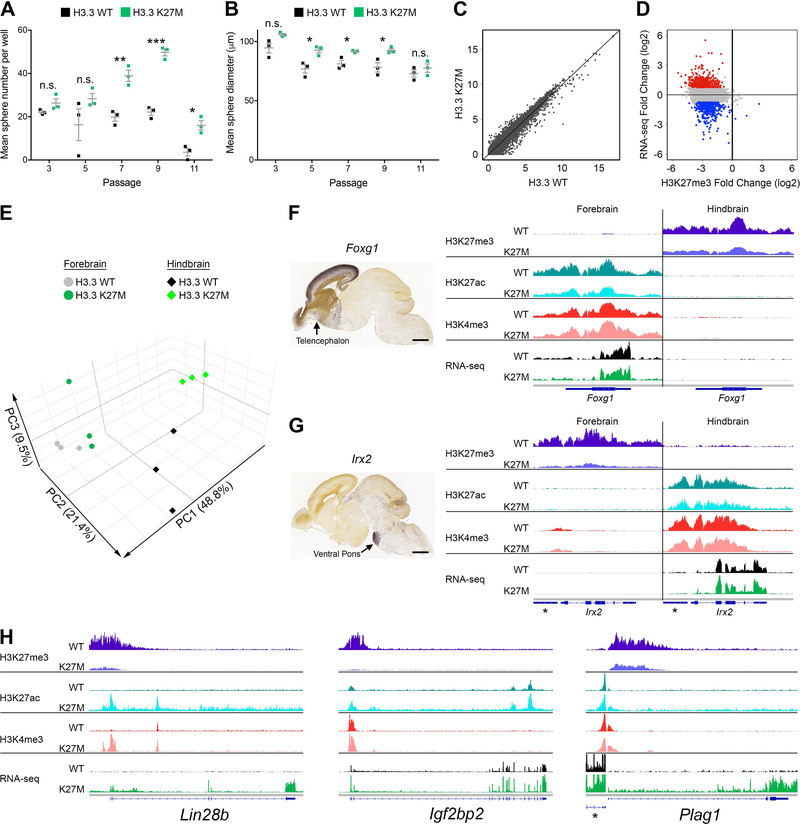
Figure 1. H3.3 K27M Promotes Self-Renewal and Mediates Global H3K27me3 Depletion but Discrete Transcription Changes That Do Not Disrupt Regional Signatures.
(A,B) Self-renewal of H3.3 K27M and H3.3 WT H-NSCs was assessed by clonogenic growth in methylcellulose at subsequent passages measuring number (A) and size (B) of spheres. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant. Error bars show +/− SEM. n = 3 per genotype (C) Scatterplot comparing expression in H3.3 K27M and H3.3 WT H-NSCs (RNA-seq; log2(FPKM+1)). (D) Plot of H3.3 K27M/H3.3 WT log2 ratio for RNA-seq versus H3K27me3 in H-NSCs. Colored dots depict genes up (red) and downregulated (blue) in H3.3 K27M compared to WT, with p < 0.05 and log2 fold change greater than 0.75 or less than −0.75, respectively, compared to the gene loci bulk (gray). (E) PCA of H3.3 WT and H3.3 K27M F- and H- NSCs. (F,G) Regional specific expression of Foxg1 (F) and Irx2 (G) shown by in situ hybridization (Allen Brain Atlas, E18.5) and average IGV tracks in H3.3 K27M F- and H-NSCs. *indicates Gm20554 locus near Irx2. Scale bar = 1 mm. (H) Average H-NSC tracks for three H3.3 K27M upregulated genes, Lin28b, Igf2bp2 and Plag1. *indicates Chchd7 locus near Plag1. In (F-H), tracks show H3K27me3, H3K27ac and H3K4me3 enrichment and RNA-seq in H3.3 WT or H3.3 K27M expressing NSCs. For each pair of tracks, n = 3 per genotype, scale is the same for both genotypes. See also Figures S1, S2, S3 and Tables S1–S3.