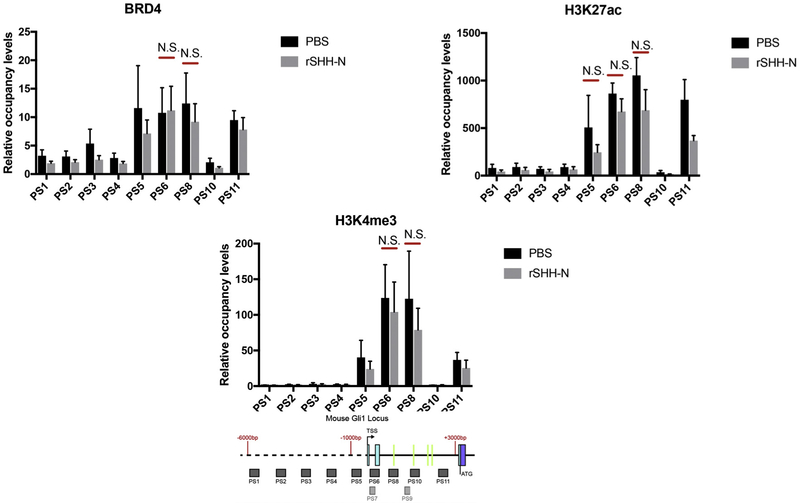
Fig. 3.
Epigenetic marks are not responsive to SHH stimulation in cells that lack GLI2 and GLI3 (Gli2−/−;Gli3−/− MEFs). Brd4, H3K27ac and H3K4me3 occupancy in the region were analyzed by ChIP-quantitative PCR in Gli2−/−;Gli3−/− MEFs treated with 1 μg/μl recombinant SHH-N (C25II Substitution) for 24 h. The signals were normalized to a control ChIP performed using rabbit IgG. Error bars represent the S.E. of three independent experiments. p values ≤ 0.05 are considered statistically significant and indicated by an asterisk. (Below) a schematic of the mouse GLI1 locus from −6,000 nt to + 3000 nt, relative to the transcription start sites (TSS) is shown. The probe sets (PS) PS1-PS11 are represented as shaded boxes (sequences in Methods section). 5′ - > 3′ is left to right. Green lines in the intron represent the locations of the mouse GBS.