Abstract
Skeletal muscle tissue engineering (SMTE) aims at repairing defective skeletal muscles. Until now, numerous developments have been done in SMTE, however it is still challenging to recapitulate the complexity of muscles with current methods of fabrication. In this review, after a brief description of the anatomy of skeletal muscle, and a short state-of-the-art on developments made in SMTE with “conventional methods”, we focus on the use of three dimensional (3D) bioprinting as a new tool for SMTE. We discuss on the current bioprinting methods, and provide an overview on the bioink formulations and properties used in 3D bioprinting. Finally, we highlight different advances made in SMTE by 3D bioprinting, and describe future needs and provide a short perspective.
Keywords: Skeletal muscle, 3D printing, Bioprinting, Bioink, Tissue engineering, Hydrogels
1. Introduction
Skeletal muscles represent ~45% of the human body weight with over 600 different skeletal muscles involved in skeletal support, stability, locomotion and dynamic events, including regulation of metabolism. They are constituted of muscle fibers wrapped by a thin connective tissue named endomysium, which are axially aligned and gathered together in a bundle covered by another protective connective tissue named perimysium.[1] Multiple bundles are arranged together and form the muscle, which is covered by the epimysium (Figure 1). Skeletal muscles are also connected to the vascular network for receipt of nutrients and waste removal, to the neuronal network for activation and contraction, and to the bones through tendons. Skeletal muscle fibers are heterogeneous and histological analysis has identified different fiber types.[2] For example, in adult muscle, fibers are often distinguished between slow (type I) fiber type and fast (type II), which have a slow or fast myosin heavy chain (MHC) isoforms correlated to a low or high actin-dependent ATP-ase. Type I fibers, which are red due to their content in myoglobin, have high mitochondrial content and generate ATP via oxidative reaction. Type II fibers are white due to the absence of myoglobin and generate ATP via glycolytic reactions. The fast muscle fibers can be subdivided in three groups: the oxidative fibers expressing MHC IIa and two glycolytic fibers with medium speed (MHC IIx) and fast speed (MHC IIb). Intermediated fibers (blended I+IIa, IIa+IIx, or IIx+IIb) also are present in muscle giving rise to a continuous spectrum of fibers.[3] These fibers are formed from the fusion of myoblasts into long, cylindrical, multi-nucleated syncytium named myotubes.[4] Under microscope, muscular (and cardiac) fibers appears striated due to the alignment in registry of repeated functional units called sarcomeres. At the intracellular level, sarcomeres are defined by the structural organization between two streaks of dense proteins, namely Z lines.[5] Between these two Z lines are the actin filaments, which are partially overlapped by myosin filaments in the central area of the sarcomere appearing under microscope as a darker area named A band.[5] When the muscle contracts the myosin filaments slide along the actin filament, overlapping each other, and the sarcomere unit decrease in size guaranteeing muscle movement.
Figure 1.
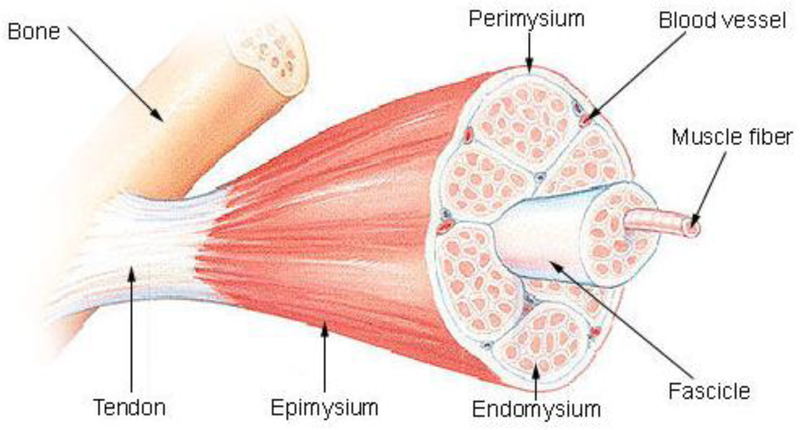
Skeletal muscle anatomy (Reproduced with permission from [1] ©2010 MedicalTerms.info)
Muscles are subjected to traumatic injury or diseases called myopathies. In the USA around 4.5 millions reconstructive surgeries are done annually resulting in billions of dollars of health care expenses.[6] Skeletal muscle tissue engineering (SMTE) aims to replace or to restore functionality in skeletal muscles that have been damaged or have lost some of their functionalities due to diseases, accidents, or severe surgeries. SMTE involves the culture of myogenic progenitor cells or stem cells obtained from the patient, the use of a scaffold in some cases or could be scaffold-free in others, and the generation of a functional skeletal muscle that can be implanted into the patient’s body.[7] SMTE has applications in regenerative medicine[8] but also in cell-based assays[9], biorobotics, biosensing, energy harvesting, and drug screening.[10–13] In this review, we highlight some standard methods to fabricate in vitro skeletal muscle tissue and their limitations. We further introduce 3D bioprinting technology and review the recent advances in the engineering of skeletal muscle tissue enabled by this technique. We finally discuss the main challenges, and future perspectives related to 3D bioprinting of skeletal muscle tissues.
2. Skeletal muscle tissue engineering
2.1. State-of-the-art
In vitro and ex vivo tissue culture studies of skeletal muscles have been established for more than a century.[14] However, to the best of our knowledge, the reconstruction of tissues from skeletal muscle cells only began as early as 1960s when Konigsberg differentiated primary chick embryonic muscle cells on petri dishes into a colony of cross-striated muscle fibers.[15] Konigsberg long pointed out the importance of the extracellular matrix (ECM) protein collagen as a critical component to the development of muscle colonies[16], which led to its widespread use in SMTE [17–19]. Since then, other natural and synthetic materials such as fibrin[20–24], alginate[25–28], polycaprolactone-based polymers[29–31] and various strategies have been developed to generate skeletal muscle tissues in vitro. Especially, the engineering of muscle fibers in vitro requires the culture of myoblasts in an anisotropic environment, promoting their alignment, favoring their fusion and the myogenesis.[32] Different methods have been developed to induce cell alignment such as the use of grooves/ridges micro-/nano-patterned substrates[33, 34], nanofibers[35, 36], anchors and hydrogel compaction, chemical surface patterning[37], stencils, mechanical stimulations, and electrical or magnetic fields.[5, 7] Moreover, to improve skeletal muscle cell differentiation and to obtain muscle tissues with high functionality, scaffolds with specific topographical features, stiffness, electrical conductivity, polymeric compositions (i.e. homopolymer, composites, hybrid nanomaterials-polymer blend) and soluble factors have been developed.[35, 36] In addition, more complex engineered tissues have been fabricated by using co-cultures of skeletal muscle cells with fibroblasts to engineer the myotendinous junction, or endothelial cells to vascularize muscle, or with neural cells to obtain neuromuscular junctions.[38] Conventional methods to fabricate skeletal muscle tissues are diverse and each one has its own merits. However, despite significant advances in SMTE, fully functional skeletal muscle tissue constructs have not yet been fabricated in vitro. In particular, the forces generated from engineered skeletal muscle tissues are still low compared to their natural counterparts[39, 40], as the in vitro muscles usually present a more immature phenotype resembling denervated muscles.[41] To improve the functionality of engineered muscles, researchers have aimed to mimic the structure and microenvironment of skeletal muscle in vivo. A common point to all conventional methods is the fabrication of anisotropic scaffolds to allow muscle cell alignment and to favor myogenesis. However, these methods have limitations in inducing precise 3D spatial cell organization. Three dimensional bioprinting techniques aim to overcome these limitations by providing high precision in cell and matrix deposition, to rapidly fabricate complex structures (Figure 2).[13]
Figure 2. Engineering skeletal muscle tissues by conventional and 3D bioprinting methods.
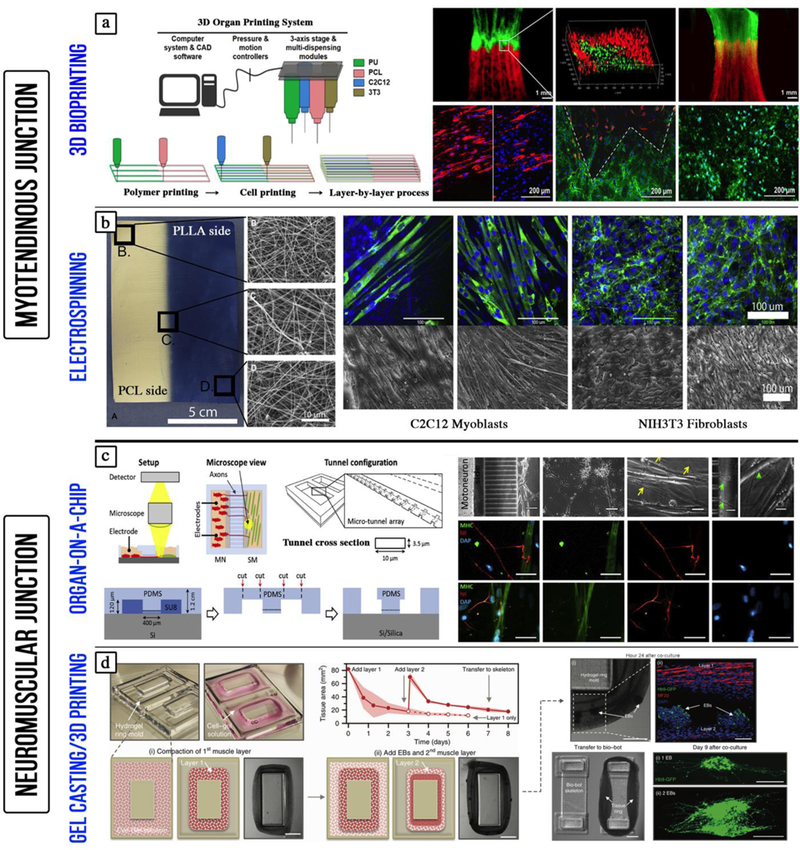
a) Engineering the myotendinous junction (MTJ) using 3D bioprinting. Myoblasts (C2C12) and fibroblasts (3T3) are precisely deposited to mimic the 3D organization of the native tissue (adapted with permission from [87] ©2015 IOP Publishing Ltd). b) Electrospun polymer gradient was used to engineer the MTJ: The mechanical stiffness increased along the gradient mimicking the interface between the tendon and skeletal muscle (adapted with permission from [135] ©2010 Elsevier Ltd). c) Engineering the neuromuscular junction (NMJ) using a microchip: The microchip allowed the co-culture of separate populations of motor neurons and human skeletal myoblasts connected through micro-channels (adapted with permission from [136] ©2018 Elsevier Ltd). d) Co-culture of muscle and embryonic bodies (EBs) of differentiated motor neurons on 3D printed hydrogel mold. The muscle-neuron tissue rings were then transferred to 3D printed bio-bot skeleton connecting two pillars (adapted with permission from [137] ©2017 Creative Commons Attribution 4.0 International License).
2.2. Bioprinting as an innovative technology to engineer skeletal muscle tissue
2.2.1. Deposition strategies
Three dimensional bioprinting refers to a set of techniques that can be used to fabricate 3D physical structures containing cells and materials. The usual techniques used in bioprinting include inkjet printing, extrusion-based printing, laser-assisted printing, and stereolithography (Figure 3).[42–49] Inkjet printing is based on a drop-by-drop bioink deposition and different operation mechanisms such as thermal, piezoelectric, electrostatic, acoustic, hydrodynamic, and microvalves are used to generate the bioink droplets.[50] The deposition of bioink droplets through the printer head is synchronized with a motorized stage allowing the fabrication of 3D constructs. Inkjet printing usually requires the use of a low viscosity bioink (<15 mPa s) and the technique is relatively cheap and fast (1–10,000 droplets per second) resulting in high cell viability (>85%).[51] Extrusion-based printing relies on pushing the bioink through a nozzle by using pneumatic or mechanical pressure. The bioink deposition, as continuous lines or small beads, occurs by raster scanning the printer head in the X-Y direction over the stages. The printer head or the stage then moves in the Z direction allowing layer-by-layer deposition. Extrusion-based printing allows the use of bioinks with a wide range of viscosities (<6×107 mPa s), it is low cost and it can be considered as a medium to fast speed (<0.05 mm/s) printing technique. It also results in high cell viability (~80%) in the printed constructs.[51] In laser-assisted printing, a ribbon in glass or quartz is coated by a thin layer of metal (i.e. gold, titanium), which is then loaded with the bioink. A laser pulse induces the vaporization of the metal film resulting in the formation of a high pressure bubble that pushes a droplet of bioink towards the substrate.[52] Laser-assisted printing requires the use of bioinks with viscosities (<300 mPa s). It is high cost and a fast speed (<1,600 mm/s) printing technique, and it results in high cell viability (95%) in the printed constructs.[51, 53] Laser-assisted printing allows high precision in bioink deposition due to its picoliter-level resolution. In stereolithography, a laser cures a photosensitive resin in a point-by-point manner to fabricate a 3D structure. However, for printing large tissues, a line-by-line writing approach by a nozzle or a laser-based bioprinters is a process that needs to be optimized. To avoid rastering the laser beam over the X-Y plane of the stage, a digital light processing (DLP) based 3D printing uses a digital micromirror array device (DMD), which consists of millions of individually addressable micromirrors that can be switched ON or OFF, to cure the whole plane of the targeted projected 3D structure in a layer by layer manner. DLP offers superior speed, resolution, scalability, and flexibility for printing 3D structure with micrometer resolution.[54] Furthermore, it avoids the formation of artificial interfaces providing better mechanical properties and a large variety of biomaterials, including nanoparticles and biomolecules, can be incorporated into the printed tissue.
Figure 3. Three major bioprinting strategies:
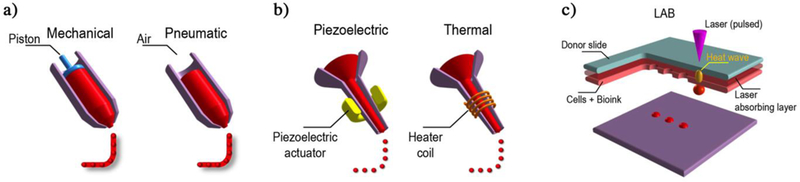
(a) Micro-extrusion printers utilize direct mechanical or pneumatic dispensing systems to extrude continuous beads of cell-containing biomaterials. (b) Inkjet bioprinters use either a local pulsed joule heater to heat the print-head and produce air bubbles forcing droplets out of the nozzle, or a piezoelectric actuator to generate localized pressure via ultrasonic waves that can form droplets of bioink-cell hybrid. (c) Laser-assisted bioprinters (LAB) emit laser beams on an absorbing substrate that generate heat waves dispensing the cell-containing materials onto the substrate.
2.2.2. Bioink formulations
Bioinks should ideally mimic the ECM of the target tissue to support cell proliferation and differentiation. Moreover, the bioink should be printable and therefore the rheological properties of the bioink are fundamental.[55, 56] Thus, bioinks with low viscosity are adapted to inkjet bioprinting, whereas more viscous bioinks are usually used in extrusion bioprinting.[57] The viscosity of the bioink and its gelling time are important for the resolution of the bioprinted structure and for preserving its shape post-printing. However, bioinks with high polymer concentration are easily printable but usually not suitable for cells since they limit the cell spreading, migration, proliferation, and matrix remodelling.[58] A trend exists in designing bioink formulations for tissue engineering applications towards the use of low polymer concentration to favour the development of tissues.[59] Additionally, a bioink should have the cells homogeneously distributed in suspension to avoid cell aggregation and deposition and to extend the bioprinting time for making larger constructs.[60, 61] The development of bioinks is an active research area, and especially for SMTE, the development of hydrogel-based bioinks is well adapted (Table 1).[24, 62–64]
Table 1.
Various bioinks formulated for engineering skeletal muscle tissue through additive manufacturing technologies.
| Bioprinting strategy | Bioink composition | Myogenic progenitors |
In vitro/In vivo results | Reference |
|---|---|---|---|---|
| FDM/DIW(ITOP) | Gelatin/fibrinogen/HA | C2C12 | • Multi-cellular constructs aimed at mimicking the myotendinous junction (MTJ) were fabricated intercalating hydrogel fibers loaded with C2C12 or NIH/3T3 between PU or PCL fibers • Zonal cellular organization typical of MTJ was recapitulated |
Merceron et al. [82] |
| Microfluidic/Co-axial bioprinting | Alginate/PEG-Fibrinogen | C2C12 | • Multi-cellular constructs with compartmentalized cell spatial organization • Anisotropic aligned structures support myoblast 3D organization and differentiation |
Costantini et al. [88] |
| Inkjet printing | Suspension of cells in sterile phosphate-buffered saline (PBS) solution | C2C12 | • High resolution (85 εm) and cell viability (>90%) • Development of bio-MEMS • Formation of confluent myotubes after 4 days on cantilevers |
Gao et al. [95] |
| SLA | Matrigel/Fibrinogen/Collagen I | C2C12 | • Maturation of myotubes after electrical stimulation • Formation of biohybrid robot powered by an antagonistic pair of skeletal muscle tissues • Inchworm-like crawling locomotion of the structure at 117.8 εm/s |
Cvetkovic et al. [96] |
| SLA | Matrigel/Fibrinogen | Optogenetic C2C12 | • Bio-bots powered by bioactuators controlled by noninvasive light stimuli • Directional locomotion (310 μm/s or 1.3 body lengths/min) and 2D rotational steering (2°/s) |
Raman et al. [98] |
| SLA | Matrigel/Fibrinogen | C2C12 | • Development of a mesoscale model for studying skeletal muscle physiology in vitro • Larger active tension forces created in muscle rings (184 ± 20 μN) after 17 days of growth in response to optical stimulation |
Raman et al. [99] |
| FDM/Electrospinning/DIW | Alginate/PEO | C2C12 | • Control of myoblast proliferation and organization by assembling a cell-laden hierarchical scaffold containing additional multi-layered PCL struts and micro/nanofibers | Yeo et al. [100], [101] |
| FDM | – | C2C12 | • Aligned scaffolds were fabricated out of PVA/PCL (3:7 ratio). After leaching PVA, samples were coated with collagen • Micropatterned/fibrous PCL bundles guided C2C12 alignment |
Kim et al. [102] |
| FDM/DIW | Muscle-derived ECM | C2C12 | • Biomimetic matrix supported higher cell viability, increased cell proliferation and myogenic gene expression compared to constructs prepared with collagen bioink. • Pre-patterning of acetylcholine receptors |
Choi et al. [103] |
| FDM/DIW | Gelatin/fibrinogen/HA/Glycerol | C2C12 | • Myotube formation after 7 days of in vitro culture • Bioprinted constructs implanted subcutaneously in nude rats showed organized muscle fibers, acetylcholine receptors, nerve contacts, and vascularization |
Kang et al. [104] |
| FDM/DIW(ITOP) | Fibrinogen/Gelatin | Human muscle progenitor cells (hMCs) isolated from biopsies | • Transplantation of the construct in a rat tibialis anterior (TA) muscle defect model • 82% restoration of the muscle force after 8 weeks post-surgery • Bioprinted constructs well-integrated with the vascular and neural networks |
Kim et al. [105] |
| 3D printing/gel casting | Silk fibroin/Collagen I/Matrigel | Human primary skeletal myoblasts (hSKMs) |
• Myoblasts can be differentiated in co-culture with NG108–15 and hiNSC-derived motoneuron-like cells in 2D and 2.5 with mature cell-specific phenotypic expression • 3D neuromuscular co-cultures can be formed with active anisotropic myofiber function. |
Dixon et al. [106] |
Natural and synthetic polymers have been used for the development of bioinks for bioprinting skeletal muscle constructs. Thus, among natural polymers, fast crosslinking hydrogels such as calcium alginate or fibrin have been used directly as bioink or as a supporting polymer during printing process in order to maintain the printed shape of less stable bioinks.[65, 66] Other natural hydrogel bioinks such as alginate, collagen, and gelatin have been widely used to provide physical support and cell supportive functionalities for engineered tissues.[67] In one example, an alginate sacrificial network template was used to entrap different pre-polymers (e.g. gelatin, agarose, gelatin methacryloyl (GelMA), polyethylene glycol diacrylate (PEGDA), polyvinyl alcohol (PVA)). These sacrificial alginate networks were first crosslinked and later removed by using a calcium chelator solution to obtain pure polymeric fibers.[68] Moreover, a patient specific bioink loaded with multiple autologous biological factors has been developed for printing 3D scaffolds by loading an alginate bioink with platelet rich plasma (PRP) used as a source of autologous growth factors for enhancing the angiogenesis, the reduction of inflammation, the stem cell recruitment, and the cardiovascular and skeletal muscle tissue regeneration.[69] To precisely tune the mechanical properties of natural polymers, various chemical functional groups such as methacryloyl groups have been conjugated to natural polymers to make them photocrosslinkable. Thus, GelMA, hyaluronic acid methacrylate (HAMA), carboxymethyl cellulose methacrylate (CMCMA), glycidyl methacrylate (GMHA), oxidized methacrylate alginate (OMA) and methacrylate alginate (MA)[70] are few examples of such polymers. This photocrosslinking allows fast crosslinking but to obtain adequate polymer crosslink the UV exposure time and photoinitiator concentration must be optimized and this will depend also on the presence of cells in the bioinks.[59, 67, 71] Additionally, photocrosslinking with visible blue light is also possible to overcome the effects of long UV light exposure time on the viability of cells.[72, 73] Thus, Bertassoni and colleagues printed cell loaded GelMA hydrogels and showed that cell-laden hydrogel constructs with different shapes, concentrations and mechanical properties could be printed, and that cells remained viable for at least 1 week in culture.[74] In another study, Liu and colleagues printed GelMA fibers at a low concentrations (<2%) by using alginate + 1% CaCl2 in the core and sheath of the nozzle to sandwich GelMA hydrogels during extrusion, allowing subsequent photocrosslinking.[75] The results showed that encapsulated cells (MCF-7, NIH 3T3, HUVECs) were viable and spread well in 1.5–2% GelMA hydrogels, whereas 1% GelMA gels were too weak to allow hydrogel formation and good cell attachment. This sandwich bioprinting strategy can be used with other hydrogels such as collagen, fibrin, and Matrigel at low concentrations. Since the delivery of nutrients to cells is a key problem in the fabrication and maintainance of thick tissues much work has been done to develop vascularized networks in muscle tissues. For example, Ma and collaborators have used a coaxial nozzle to fabricate hollow constructs and showed higher cell viability in the gel channels compared to those made in PDMS channels.[76] Also, a vascularized cardiac tissue was made by bioprinting endothelial cells and neonatal cardiomyocytes in composite GelMA (3.5–5%)-alginate (4%) bioink. Later, they integrated this engineered tissue into a microbioreactor and showed its application in drug testing by studying the effects of doxorubicin on the beating rate of the myocardium tissue.[67] In another study, a bioink composed of GelMA (5–7%), sodium alginate (1–3%), and 4-arms poly(ethylene glycol) tetra-acrylate (PEGTA) (1–3%) was used to bioprint endothelial cells (HUVECs) and mesenchymal stem cells (MSCs), which in presence of TGF-β1 differentiated into smooth muscle cells (SMCs). Their results showed that both cells proliferated and differentiated as shown by the expression of CD31 for endothelial cells and α-smooth muscle actin (α-SMA) for SMCs.[71] To induce tissue formation, decellularized extracellular matrix (dECM) has also been used. Indeed, dECM matrices are tissue specific, and since they are extracted from the tissue itself, they are the most biomimetic materials and contain a variety of proteins, proteoglycans and cytokines, that can aid in directed differentiation of stem cells, tissue formation and maturation.[77, 78] To date, dECM-based bioinks have been prepared from different tissues including liver, heart, cartilage, skin, vasculature, brain, lung, kidney, bone, spinal cord, colon, umbilical cord, pancreas, adipose tissue, skeletal muscle and have been used in most of the cases to regenerate the same tissue of origin.[79–81] However, dECM-based bioinks form gels with low mechanical properties and may require stiffening by the use of crosslinking agents. To overcome this problem, dECM-based bioinks have also been blended with different components such as gelatin, PEG derivatives, and PCL to improve their viscosity, mechanical properties, and printability.[82, 83] Despite promising results, the drawbacks of dECM-based bioinks are the batch-to-batch variability and the potential immune responses they may induce in vivo upon implantation.
Among synthetic polymers, PEG-based hydrogels have been extensively used for bioprinting. However, other polymers including PLGA, PCL, or PVA are also frequently employed.[84, 85] Furthermore, methacrylate and acrylate functional groups have been conjugated to synthetic polymers to make them photocrosslinkable. Thus, PEGDA, poly(ethylene glycol) dimethacrylate (PEGDMA), star poly(ethylene glycol-co-lactide) acrylate (SPELA), poly(ethylene oxide) dimethacrylate (PEODMA) are some examples of these modified polymers.[86] Among synthetic polymers, polyurethane (PU) has also been widely used for medical applications due to its good mechanical strength, flexibility, hydrophobicity and biocompatibility. In one example, a thermoplastic PU construct was loaded with C2C12 and a PCL construct was loaded with fibroblasts for engineering a muscle-tendon tissue interface.[87] However, PU is usually synthesized using isocyanates, which is derived from a reaction between amine and phosgene. The residual, toxic unreacted products and derived aromatic diamine products leaking from PU are of concern in vivo. To overcome this problem, several groups have used green chemistry to produce PU via a phosgene and isocyanate free synthesis pathway.[88]
Composite bioinks of natural-natural, synthetic-synthetic, and natural-synthetic polymers, take advantage of the cell-supportive properties from the natural polymer and the mechanical properties and tunability of the synthetic polymer, making them ideal for muscle tissue engineering via bioprinting. In one study, Alsberg and collaborators fabricated an OMA-PEG hydrogel for bioadhesive and tissue engineering applications via bioprinting.[89] Poly(vinyl alcohol)(PVA)-alginate has also been used to bioprint constructs with micropores showing controlled release of proteins.[90] In another study, Ramon-Azcon and colleagues developed a library of composite biomaterials used as bioinks, and showed that GelMA-CMCA allowed the growth and differentiation of encapsulated C2C12, while presenting high resistance to degradation.[91] Moreover, microfluidic print heads have also been used to deposit multiple materials into microfibers or droplets. This technique allows fast switching between different bioinks as well as the formation of fibers with different coded bioink segments or heterogeneous fibers with two or more parallel bioinks in the fiber.[59, 92] Thus, Rainer and collaborators used a microfluidic head to print two different cell types, namely C2C12 and BALB/3T3 fibroblasts, in a Janus fiber employing PEG-fibrinogen/alginate bioink. They observed high cell compartmentalization in each half of the extruded fiber. After 5 days of culture, good spatial organization with myotube formation in the C2C12 printed half and fibroblast compartmentalization in the opposite side was observed.[93] To avoid cell deposition in the ink and to allow longer bioprinting, materials that act as surfactants have also been added into the inks. Thus, Panhuis and colleagues[61] reported the development of a novel bioink by using drop-on-demand printing. The system uses both a commercial micro-valve deposition system and multi-nozzle piezoelectric inkjet print heads to allow facile cell deposition. The novel bioink is composed of gellan gum in Dubelcco’s Modified Eagles Medium (DMEM) mixed with Poloxamer 188 surfactant and it can be used to reproducibly print multiple cell types over long printing periods without cell sedimentation in the ink. New bioink development exploits stimuli-responsive polymers to tailor the properties of the inks to obtain smart bioinks. Thermoresponsive polymers such as poly(N-isopropylacrylamide) (pNIPAAM) have been used in bioinks because of their lower critical solution temperature (LCST 32°C) that allow the phase transition from liquid (<32°C ) to gel (>32°C ).[94] This reversible and fast gelling characteristic allows loading of cell-laden ink in its liquid state in the printer cartridge, whereas the rapid gelation right after deposition onto a heated substrate maintains the shape of the printed constructs until the next crosslinking step.[63, 95, 96] Other smart bioinks have been made with shear-thinning property, therefore their viscosity decreases under increasing shear rate, allowing high printing fidelity since the polymer deposition is easier under pressure in the nozzle. The use of shear thinning materials results in fast gelation after the pressure is released.[97] Thus, Burdick and collaborators directly printed a shear-thinning hydrogel (25% hyaluronic acid modified with adamantine or β-cyclodextrin) into a self-healing hydrogel (40% hyaluronic acid modified with adamantine or β-cyclodextrin) that facilitated high precision printing by using the self-healing property to maintain the shape of the printed construct.[98] A recent study, by Popov and colleagues, summarized the literature over 15 years on the most popular inks used in 3D printing of skeletal muscle (Figure 4).[99]
Figure 4.
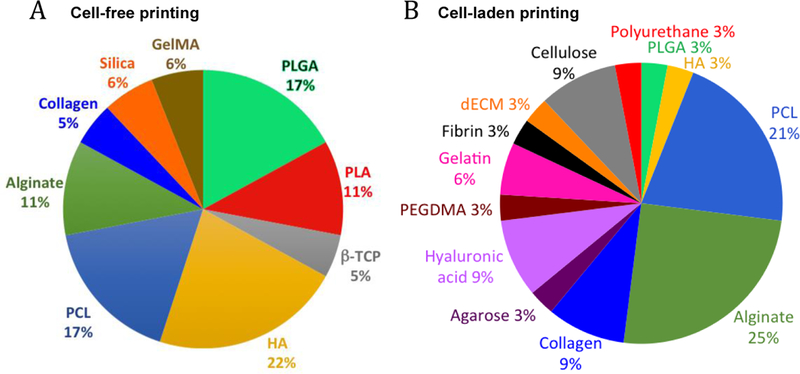
Percentage of materials used in skeletal muscle 3D printing over a period of 15 years from published articles. A) Cell-free and B) Cell-laden constructs. Adapted with permission from [99] ©2017 Future Medicine Ltd
2.2.3. Three dimensional printing and bioprinting in skeletal muscle tissue engineering
Three dimensional bioprinting allows precise deposition of matrix and cells for the fabrication of complex structures. For example, Gao and Cui reported that bioprinting can be used to precisely deposit mouse myoblasts (C2C12) in a matrix on cantilevers with a resolution of 85 μm, >90% cell viability, and with high reproducibility.[100] After differentiation, the myotubes on the cantilevers became excitable (2V, 40ms, 5Hz). Their results led them to conclude that bioprinting muscle cells in biological microelectromechanical systems (bio-MEMS) induces better physiological responses due to precise cell positioning and alignment compared to hand-based or syringe-based cell seeding where cells are seeded randomly. Cardiac cells allow taking advantage of their spontaneous beating to fabricate biological actuators. However, the development of bioactuators that responds to stimuli with controlled movements requires the use of skeletal muscle cells. Bashir and colleagues fabricated by stereolithography a biological device (bio-bots) composed of two stiff pillars with different lengths connected by a compliant beam. C2C12, ECM proteins, and Matrigel in solution were cast around and between the pillars to form a cell strip by gelation (Figure 5a). After differentiation, compaction of the gel and tension induced between the two pillars favor the maturation of myotubes which, under electrical pulses at 1 Hz, contracted and induced an inchworm-like crawling locomotion of the structure at 117.8 μm/s.[101] Advantageously, the use of stereolithographic 3D printing enhanced the number of materials and cell types, which can be used for the development of biological machines. Recently, to overcome the limitation due to spontaneous shrinkage of skeletal muscle tissue which contracts on flexible substrate, a biohybrid robot powered by an antagonistic pair of skeletal muscle tissues has been fabricated. Such robot was composed of a 3D-printed resin skeleton bearing electrodes for the active stimulation of myoblast-laden hydrogel sheets, which were mounted on two sides of the skeleton to act as antagonist muscles (Figure 5b). The authors showed that the biohybrid robot was capable of large movements (rotation angle of the joint was close to 90°) that can be used to perform simple actions (e.g. grabbing and transporting small objects).[102] Furthermore, they showed that the hybrid robot was successfully actuated over a long period of time (~1 week). Despite much success, this research field is still at its infancy and additional work will be needed to develop reliable hybrid robots for advanced applications. However, actuation by electrical stimulations has some disadvantages such as the coupling of the actuator to the surrounding environment, and the possible formation of bubbles by electrolysis that will damage the skeletal muscle tissue and the electrode. To overcome these limitations, neural stimulations can be used by co-culturing skeletal muscle cells with motor neurons, whereas skeletal muscle cells have also been genetically modified to respond to optical stimulations. Thus, Bashir and colleagues developed biological actuators powered by optogenetic skeletal muscle. These muscles were able to generate up to a 300 μN force in response to an optical stimulus. Moreover, they made damages to the skeletal tissue by transverse laceration and they developed a method to heal high volume damage in muscle bioactuators based on new myoblasts, ECM proteins, exercises, and local release of insulin-like growth factor (IGF-1) from a biological glue.[103, 104] A different approach that is becoming increasingly popular consists of exploiting the advantages of additive manufacturing, both with or without other scaffold fabrication technologies, to assemble advanced constructs that recapitulate skeletal muscle tissue organization and function. Some studies have focused on the 3D fabrication of a group of muscle fibers forming a fascicle. Thus, Yeo and Kim fabricated bundles of aligned and random polycaprolactone (PCL) microfibers using wet electrospinning. To obtain the alignment of the microfibers they stretched them at 45–50 °C. To further mimic natural muscle, a second group of scaffolds with aligned microfibers was fabricated and the microfibers were coated with collagen. To obtain an homogeneous cell seeding, they bioprinted C2C12 in 2% collagen-2% poly(ethylene oxide) (PEO) on the different scaffolds. They observed higher sarcomeric organization and differentiation at day 7 of culture on collagen coated aligned fibers and aligned fiber scaffolds compared to random fiber scaffolds.[105, 106] In another example of biomimetic muscle bundle fabrication, Kim and colleagues fabricated a microfibrous PCL bundle by using a melt-printing system to print at 85 °C a PVA/PCL (ratio 3:7) solution with a 350 μm nozzle at a speed of 10 mm/s and a pneumatic pressure of 250 kPa. The sacrificial PVA was removed in water after 24 h, then the PCL structure was coated with 0.5% collagen crosslinked by EDC for 30 min, and the scaffold was freeze dried for 12 h. Due to the microfibrillation of PVA and the leaching of PVA from the mixture of PVA/PC, the scaffold had a surface with aligned microfibrous pattern and a section allowing cell penetration between the microfibers (Figure 6a). Myoblasts (C2C12) were seeded on these scaffolds and cultured for two weeks. Analysis of cells showed longitudinal cell alignment, good cell proliferation on the surface, high cell infiltration between the microfibers, and resulted in a scaffold section mimicking a muscle bundle section. Myosin heavy chain (MHC) was also well developed.[107] Furthermore, different bioinks have been used to promote cell survival, printability, and tissue formation. Interface tissue engineering (ITE) requires materials with different mechanical and chemical properties specific to the cell types in the different areas. To engineer a muscle-tendon unit, Atala and collaborators used a composite hydrogel bioink made of gelatin, fibrinogen, hyaluronic acid, and calcium free high glucose Dulbecco Modified Eagle Medium (DMEM) to bioprint C2C12 on a printed polyurethane aligned-fiber scaffold that mimics muscle’s elasticity, and to bioprint NIH/3T3 cells on a printed PCL aligned-fiber scaffold that mimics tendon stiffness, whereas the interface area was built by overlapping (10%) the two fiber types. The constructs were then crosslinked for 30 min in 20 U/mL thrombin solution with 0.5 mM CaCl2 and put in an incubator with culture medium. After 1 day of culture, the culture medium was changed for a differentiation medium for 7 days, and the constructs were analyzed. Over 94% cell viability was observed after printing and at day 7 of culture, C2C12 expressed desmin and MHC, and fibroblasts secreted type I collagen. At the interfacial region, composed of an overlapped PU-PCL with both cell types, a distinct secretion pattern between the muscle and tendon region was observed.[87] This study showed the versatility of the integrated organ printing (IOP) used to fabricate complex constructs with region specific mechanical and biological characteristics. In another study with the IOP, Atala and collaborators fabricated human scale tissue constructs such as the mandible, calvarial bone, ear cartilage, and skeletal muscle (Figure 6c). They bioprinted C2C12 using a bioink composed of gelatin, fibrinogen, hyaluronic acid (HA), and glycerol. They observed 97% cell viability after printing, cell alignment at day 3 of culture, and myotube formation after 7 days in differentiation medium. The constructs were implanted subcutaneously in nude rats with the dissected common peroneal nerve (CPN) inserted in the constructs. After 2 weeks, nerve integration in the constructs was observed with the presence of acetylcholine receptors (AChR) clusters on the muscle fibers, and nerve contacts. Moreover, the vascularization of the constructs was induced as shown by the expression of endothelial cell markers, while electromyography showed that the engineered muscles respond to electrical stimulations and were still immature.[108] To increase cell signaling, some groups have proposed the use of bioinks composed of decellularized matrix from skeletal muscle. For example, Cho and colleagues prepared a decellularized bioink (mdECM) from porcine skeletal muscle and used it at 1% concentration to print different patterns (parallel lines with 500 μm width, diamonds, chains) of C2C12 at 18 °C (Figure 6b). After gelation at 37 °C and culture of the constructs for 1, 4 and 7 days, they observed high cell viability and increased cell proliferation compared to similar constructs prepared with collagen bioink. After induction of cell differentiation, higher myogenic gene expression was observed at day 14 of culture in C2C12 encapsulated in mdECM constructs compared to collagen constructs. Furthermore, it was observed that in addition to retain major ECM components such as laminin, collagen and glycosaminoglycans (GAGs), mdECM preserved also agrin, which allowed the pre-patterning of acetylcholine receptors (AChRs).[109] In another study, Lee and collaborators used the ITOP system to engineer a skeletal muscle construct (10×7×3 mm3) by bioprinting human muscle progenitor cells (hMCs) isolated from biopsies in a fibrin bioink associated with gelatin and PCL deposition. They transplanted the construct in a rat tibialis anterior (TA) muscle defect model and observed enhancement of the tetanic force and TA muscle weight at 4 and 8 weeks post-implantation. Moreover, 82% of the muscle force was restored after 8 weeks post-surgery compared with normal TA muscle.[110] In addition, the construct was well-integrated with the vascular and neural networks as confirmed by immunostaining and the observation of new blood vessels and mature neuromuscular junction (NMJ). In another study, Kaplan and colleagues printed 40% (w/v) silk fibroin T-shape cantilevers in 12 well plate and used them as anchors for culturing primary human myoblast-laden silk (1%)-collagen type I (3 mg/mL) -Matrigel (8%) hydrogel. After 3 days of culture, the growth medium was replaced by the differentiation medium. After 21 days the formed myotubes were characterized. In parallel, human induced neural stem cells (hiNSCs) derived motorneuron-like cells were differentiated for 2 weeks showing neurite extensions and were seeded on the differentiated myoblasts and the co-culture constructs was kept in motorneuron differentiation medium for 2 weeks. Functional neuromuscular junctions (NMJ) were observed by stimulation of calcium transients with 1.5 mM L-glutamic acid and their suppression by glutamic blockade via turbocurarine.[111] Furthermore, microfluidics has been combined with 3D bioprinting to fabricate multi-material constructs and other complex structures for SMTE. A proof-of-concept has been recently reported by Rainer and collaborators (Figure 6d). In this study, a microfluidic printing head has been used to precisely compartmentalize two different photocurable bioinks (PEG-fibrinogen, and PEG-fibrinogen-alginate), each loaded respectively with C2C12 and BALB/3T3 fibroblasts, within bioprinted hydrogel struts. Janus-like compartmentalization of the two cell types remained after 5 days of culture. In vitro analysis at day 21 of culture showed aligned, multinucleated, fully striated myotubes with abundant myosin heavy chain (MHC).[93] Moreover, after 7 d in vitro culture the constructs were implanted subcutaneously in mice and were retrieved after 28 d. The analysis showed complete maturation of tightly-packed fully striated myotubes.
Figure 5. Engineering biohybrid robots.
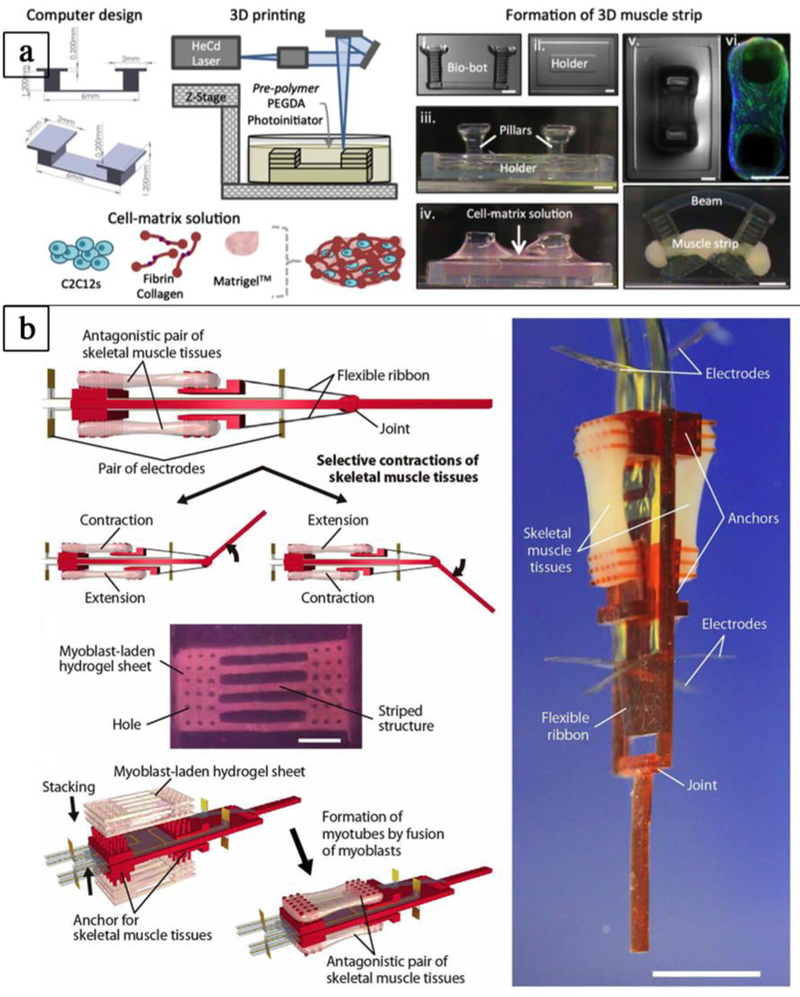
a-b) Assembling of biohybrid robots combining rapid prototyping techniques and living cells. a) 3D printed hydrogel “bio-bots” with an asymmetric physical design and powered by the actuation of an engineered mammalian skeletal muscle strip (adapted with permission from [101] ©2014 PNAS). b) Biohybrid robot powered by an antagonistic pair of skeletal muscle tissues (adapted with permission from Morimoto et al.[102] ©2018 American Association for the Advancement of Science).
Figure 6. Additive manufacturing in skeletal muscle tissue engineering.
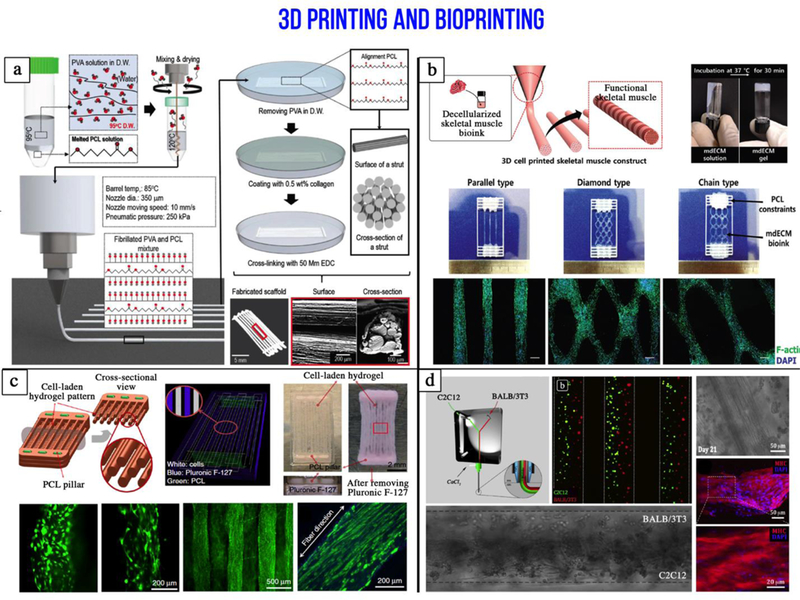
a) Fabrication processes and optical/SEM images of the hybrid microfibrillated PCL/collagen scaffold used to mimic skeletal muscle hierarchical organization (adapted with permission from [107] ©2018 John Wiley and Sons). b) Three dimensional printing of an ink made from a decellularized (mdECM) porcine skeletal muscle to promote myoblast differentiation (adapted with permission from [109] ©2016 John Wiley and Sons). c) Integrated tissue-organ printer (ITOP) to fabricate human scale tissue constructs. Muscle precursors are encapsulated within hydrogel fibers that are supported by PCL pillars (adapted with permission from [108] ©2016 Springer Nature). d) Microfluidic printing head used to precisely deposit heterogeneous janus-like hydrogel fibers containing myoblasts and fibroblasts (adapted with permission from [93] ©2017 Elsevier).
3. Conclusion and perspectives
Three dimensional bioprinting technologies (inkjet bioprinting, extrusion bioprinting, laser-based bioprinting) have great potential in tissue engineering, regenerative medicine, and drug development. These technologies should help overcome the shortage of organs for transplants, whereas the opportunity to fabricate healthy or diseased tissue models has the potential to improve the accuracy and safety of drug screening, and our understanding of developmental biology. In the past decade, these technologies have evolved and become more sophisticated and some human anatomical parts (ears, nose, vagina, and hydroxyapatite bone substitutes) using bioprinting have already been used in the clinic.[112–114] However, each printing process has its own advantages and limitations, and, as the needs for more complex tissue fabrication increases, many challenges still remain. In particular, progress in the bioprinting of soft materials (e.g. hydrogels) has not evolved as quickly as the printing of hard materials.[115] One aspect that will certainly require further improvement is printing resolution. This feature is fundamental as cells should be deposited ideally with a resolution comparable to their size (5–10 μm) to closely mimic their organization in native tissue. So far, printing resolution of available technologies is often one order of magnitude higher than the cell size, thus limiting the fabrication of advanced constructs and their functionalities.[116, 117] Other technological challenges that should be improved are the speed and reproducibility of the printing process to develop high-throughput 3D bioprinting.[115, 118, 119]. Current printing speed is generally adequate for bioprinting small scale tissue models (scaffold volume generally less than 1 cm3) that are suitable for in vitro studies. To reach the clinic, the throughput must be increased by at least one order of magnitude. Moreover, the bioprinting of thick skeletal muscle tissue (> 1 mm) is still challenging due to the requirement of an incorporated vascular network for oxygen delivery and waste removal.
Moreover, further developments are needed in the bioprinting of human cells including cells derived from patients, iPS cells, and stem cells which will allow the development of personalized medicine with patient specific implants or drugs.[119] Cell sources that can be expanded or harvested in large quantities (hundreds of millions) must be identified. In the specific case of skeletal muscle tissue, this is not trivial. Many cell sources are currently available but most of them have limited capacity to be expanded in vitro (as in the case of satellites cells). To date, one of the best candidates are pericytes – perivascular cells[120] – that can be isolated from different tissues. Skeletal muscle pericytes (MPs) showed multipotent capability and are able to differentiate into different mesodermal cell lineages such as adipocytes, smooth muscle cells, and skeletal muscle cells in relation to various stimuli of differentiation. Another suitable candidate is the induced pluripotent stem cells (iPSCs). These cells can be derived from a patient’s own cells harvested from a non-invasive skin biopsy. They possess unlimited proliferation capacity and can be differentiated into any type of tissue, including skeletal muscle tissue. Despite the great potential of iPSCs, differentiation and purification protocols represent an ongoing challenge that currently limits their translation into the clinical. Human pluripotent stem cells (hPSCs) can be differentiated into skeletal muscle cells via small molecules during differentiation or by direct reprogramming to induce overexpression of transcription factors Pax 7 and MyoD.[121–124] In a recent paper, Bursac and colleagues developed a protocol using GSK3 inhibitor CHIR99021(10 μM) followed by induction of the satellite cell marker Pax 7 to derive hPSCs into expandable myogenic progenitors, that differentiate into skeletal muscle cells and myotubes.[125]
Another major bioprinting hurdle is in the development of bioinks. Indeed, materials used in 3D bioprinting should be extrudable, crosslinkable, biocompatible, support cell growth and differentiation, and retain their shape for a certain period of time in a variety of environmental conditions. Furthermore in SMTE, the gelled bioink should have comparable mechanical properties to that of skeletal muscle tissue. The fabrication process should be balanced between the requirements for making robust biopolymer constructs and for achieving biologically relevant cell densities. In the future, the use of stimuli-responsive hydrogels in bioprinting may contribute to the development of smart bioinks.[126] Furthermore, despite being reported in different studies, multimaterial and multicellular bioprinting has been scarcely explored due to the complexity of the printer set-up that generally does not allow precise cell compartmentalization.[127] Natural tissues, such as skeletal muscle tissue, have complex multicellular anisotropic structure in relation with the nervous and vascular networks. Such complexity can be captured via the use of more complex bioprinting processes combining different techniques, bioinks, and cell types.[115] For example, the integration of microfluidics with bioprinting has allowed the fabrication of gradient structures.[59, 92, 128, 129] The merging of different bioinks has allowed the combination of different cell types since each bioink should specifically mimic the ECM of the intended tissue type. Moreover, since the spatial and temporal composition of the ECM change during cellular proliferation and differentiation, the bioinks should also reflect this dynamic evolution. Direct in situ bioprinting on or in patients is also a key future growth area with applications in medical robotics and computer-assisted medical interventions.[117] [130] Some studies have shown the feasibility of this approach. Recently, Soker and colleagues printed stem cells derived from amniotic fluid in fibrin-collagen ink on large skin wounds in mice. They showed faster wound closure, improved vascularization and skin regeneration.[131] In another study, researchers have also bioprinted nano-hydroxyapatite into mouse calvaria defects.[132] At least, common standards for additive manufacturing technologies should be established to ensure quality control at each step of the manufacturing process to allow fast translation of biofabricated skeletal muscle tissues for clinical applications.[117, 133, 134]
Acknowledgments
The authors acknowledge funding from the National Institutes of Health AR073135, RO1AR057837, and EB021857–01A1. This study was also partly financially supported by National Centre for Research and Development in the framework of the project “Consolidation of 3D printing, cell biology and material technology for the development of bioprinted meat – A prototype study” (grant no PL-TWIII/5/2016) and project “START” (grant no STRATEGMED1/233224/10/NCBR/2014). Furthermore, this work was supported by the German Research Foundation (DFG) within the collaborative research centre TRR225 (subproject B03).
References
- [1].M. T. E. O. GenericLook.com Skeletal muscle anatomy. http://medicalterms.info/anatomy/Skeletal-Muscles/.
- [2].Bottinelli R, Reggiani C, Prog. Biophys. Mol. Biol. 2000, 73, 195. [DOI] [PubMed] [Google Scholar]
- [3].Liu Y, Shen T, Randall WR, Schneider MF, J. Muscle Res. Cell Motil. 2005, 26, 13. [DOI] [PubMed] [Google Scholar]
- [4].Sanger JW, Wang J, Fan Y, White J, Sanger JM, J. Biomed. Biotechnol. 2010. 2010, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ostrovidov S, Hosseini V, Ahadian S, Fujie T, Parthiban SP, Ramalingam M, Bae H, Kaji H, Khademhosseini A, Tissue Eng. Part B Rev. 2014, 20, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grasman JM, Zayas MJ, Page RL, Pins GD, Acta Biomater. 2015, 25, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ostrovidov S, Shi X, Sadeghian R, Salehi S, Fujie T, Bae H, Ramalingam M, Khademhosseini A, Stem Cell Rev. Rep. 2015, 1. [DOI] [PubMed] [Google Scholar]
- [8].Hutmacher D, Duda G, Guldberg R, Cell Tissue Res. 2012, 347, 485. [DOI] [PubMed] [Google Scholar]
- [9].Nagamine K, Kawashima T, Sekine S, Ido Y, Kanzaki M, Nishizawa M, Lab Chip 2011, 11, 513. [DOI] [PubMed] [Google Scholar]
- [10].Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A, Drug Discov. Today 2012, 17, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vandenburgh H, Tissue Eng. Part B Rev. 2009, 16, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Neal D, Sakar MS, Bashir R, Chan V, Asada HH, Tissue Eng. Part A 2015, 21, 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cui X, Gao G, Qiu Y, Biotechnol. Lett. 2013, 35, 315. [DOI] [PubMed] [Google Scholar]
- [14].Lewis MR, Am. J. Physiol. 1915, 38, 153. [Google Scholar]
- [15].Konigsberg IR, Proc. Natl. Acad. Sci. U.S.A. 1961, 47, 1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hauschka SD, Konigsberg IR, Proc. Natl. Acad. Sci. U.S.A. 1966, 55, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Beier JP, Klumpp D, Rudisile M, Dersch R, Wendorff JH, Bleiziffer O, Arkudas A, Polykandriotis E, Horch RE, Kneser U, BMC Biotechnol. 2009, 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kroehne V, Heschel I, Schügner F, Lasrich D, Bartsch JW, Jockusch H, J. Cell. Mol. Med. 2008, 12, 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Osaki T, Sivathanu V, Kamm RD, Biomaterials 2018, 156, 65. [DOI] [PubMed] [Google Scholar]
- [20].Huang Y-C, Dennis RG, Larkin L, Baar K, J. Appl. Physiol. 2005, 98, 706. [DOI] [PubMed] [Google Scholar]
- [21].Nagamine K, Kawashima T, Ishibashi T, Kaji H, Kanzaki M, Nishizawa M, Biotechnol. Bioeng. 2010, 105, 1161. [DOI] [PubMed] [Google Scholar]
- [22].Matthias N, Hunt SD, Wu J, Lo J, Smith Callahan LA, Li Y, Huard J, Darabi R, Stem Cell Res. 2018, 27, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pollot Beth E, Rathbone Christopher R, Wenke Joseph C, Guda T, J. Biomed. Mater. Res. B: Appl. Biomater. 2017, 106, 672. [DOI] [PubMed] [Google Scholar]
- [24].Lev R, Seliktar D, J. Royal Soc. Interface 2018, 15, 20170380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rowley JA, Madlambayan G, Mooney DJ, Biomaterials 1999, 20, 45. [DOI] [PubMed] [Google Scholar]
- [26].Borselli C, Storrie H, Benesch-Lee F, Shvartsman D, Cezar C, Lichtman JW, Vandenburgh HH, Mooney DJ, Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schuster E, Wallin P, Klose FP, Gold J, Ström A, Food Hydrocoll. 2017, 72, 210. [Google Scholar]
- [28].Ansari S, Chen C, Xu X, Annabi N, Zadeh HH, Wu BM, Khademhosseini A, Shi S, Moshaverinia A, Ann. Biomed. Eng. 2016, 44, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fasolino I, Guarino V, Cirillo V, Ambrosio L, J. Biomed. Mater. Res. A 2017, 105, 2551. [DOI] [PubMed] [Google Scholar]
- [30].Choi JS, Lee SJ, Christ GJ, Atala A, Yoo JJ, Biomaterials 2008, 29, 2899. [DOI] [PubMed] [Google Scholar]
- [31].Kim Min S, Jun I, Shin Young M, Jang W, Kim Sun I, Shin H, Macromol. Biosci. 2009, 10, 91. [DOI] [PubMed] [Google Scholar]
- [32].Jana S, Levengood SKL, Zhang M, Adv. Mater. 2016, 28, 10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shi X, Ostrovidov S, Zhao Y, Liang X, Kasuya M, Kurihara K, Nakajima K, Bae H, Wu H, Khademhosseini A, Adv. Funct. Mater. 2015, 25, 2250. [Google Scholar]
- [34].Gao H, Cao X, Dong H, Fu X, Wang Y, J. Mater. Sci. Technol. 2016, 32, 901. [Google Scholar]
- [35].Ostrovidov S, Shi X, Zhang L, Liang X, Kim S, Fujie T, Ramalingam M, Chen M, Nakajima K, Al-Hazmi F, Bae H, Memic A, Khademhosseini A, Biomaterials 2014, 35, 6268. [DOI] [PubMed] [Google Scholar]
- [36].Ostrovidov S, Ebrahimi M, Bae H, Nguyen HK, Salehi S, Kim SB, Kumatani A, Matsue T, Shi X, Nakajima K, Hidema S, Osanai M, Khademhosseini A, ACS Appl. Mater. Interfaces 2017, 9, 42444. [DOI] [PubMed] [Google Scholar]
- [37].Fujie T, Ahadian S, Liu H, Chang H, Ostrovidov S, Wu H, Bae H, Nakajima K, Kaji H, Khademhosseini A, Nano Lett. 2013, 13, 3185. [DOI] [PubMed] [Google Scholar]
- [38].Ostrovidov S, Ahadian S, Ramon-Azcon J, Hosseini V, Fujie T, Parthiban SP, Shiku H, Matsue T, Kaji H, Ramalingam M, Bae H, Khademhosseini A, J. Tissue Eng. Regen. Med. 2014, 11, 582. [DOI] [PubMed] [Google Scholar]
- [39].Uzel SGM, Pavesi A, Kamm RD, Prog. Biophys. Mol. Biol. 2014, 115, 279. [DOI] [PubMed] [Google Scholar]
- [40].Dennis RG, Kosnik PE, Gilbert ME, Faulkner JA, Am. J. Physiol. Cell Physiol. 2001, 280, C288. [DOI] [PubMed] [Google Scholar]
- [41].Cheng CS, Davis BNJ, Madden L, Bursac N, Truskey GA, Exp. Biol. Med. 2014, 239, 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nakamura M, Kobayashi A, Takagi F, Watanabe A, Hiruma Y, Ohuchi K, Iwasaki Y, Horie M, Morita I, Takatani S, Tissue Eng. Pt A 2005, 11, 1658. [DOI] [PubMed] [Google Scholar]
- [43].Boland T, Xu T, Damon B, Cui X, Biotechnol. J. 2006, 1, 910. [DOI] [PubMed] [Google Scholar]
- [44].Khalil S, Sun W, Mater. Sci. Eng. C 2007, 27, 469. [Google Scholar]
- [45].Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Rémy M, Bordenave L, Amédée J, Guillemot F, Biomaterials 2010, 31, 7250. [DOI] [PubMed] [Google Scholar]
- [46].Gaebel R, Ma N, Liu J, Guan J, Koch L, Klopsch C, Gruene M, Toelk A, Wang W, Mark P, Wang F, Chichkov B, Li W, Steinhoff G, Biomaterials 2011, 32, 9218. [DOI] [PubMed] [Google Scholar]
- [47].Gauvin R, Chen Y-C, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A, Biomaterials 2012, 33, 3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Soman P, Chung Peter H, Zhang AP, Chen S, Biotechnol. Bioeng. 2013, 110, 3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gudapati H, Dey M, Ozbolat I, Biomaterials 2016, 102, 20. [DOI] [PubMed] [Google Scholar]
- [51].Jian H, Wang M, Wang S, Wang A, Bai S, Bio-Design and Manufacturing 2018, 1, 45. [Google Scholar]
- [52].Keriquel V, Oliveira H, Rémy M, Ziane S, Delmond S, Rousseau B, Rey S, Catros S, Amédée J, Guillemot F, Fricain J-C, Sci. Rep. 2017, 7, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nathan RS, David TC, Yong H, Nurazhani Abdul R, Yubing X, Douglas BC, Biofabrication 2010, 2, 032001. [Google Scholar]
- [54].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S, Biomaterials 2017, 124, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Billiet T, Vandenhaute M, Schelfhout J, Vlierberghe S, Dubruel P, Biomaterials 2012, 33. [DOI] [PubMed] [Google Scholar]
- [56].Chung JHY, Naficy S, Yue Z, Kapsa R, Quigley A, Moulton SE, Wallace GG, Biomater. Sci. 2013, 1, 763. [DOI] [PubMed] [Google Scholar]
- [57].Murphy SV, Skardal A, Atala A, J. Biomed. Mater. Res. Part A 2013, 101A, 272. [DOI] [PubMed] [Google Scholar]
- [58].Holzl K, Lin S, Tytgat L, Van Vlierberghe S, Gu L, Ovsianikov A, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [59].Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A, Adv. Mater. 2016, 28, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hoch E, Hirth T, Tovar GEM, Borchers K, J. Mater. Chem. B 2013, 1, 5675. [DOI] [PubMed] [Google Scholar]
- [61].Ferris CJ, Gilmore KJ, Beirne S, McCallum D, Wallace GG, in het Panhuis M, Biomater. Sci. 2013, 1, 224. [DOI] [PubMed] [Google Scholar]
- [62].Pescosolido L, Vermonden T, Malda J, Censi R, Dhert WJA, Alhaique F, Hennink WE, Matricardi P, Acta Biomater. 2011, 7, 1627. [DOI] [PubMed] [Google Scholar]
- [63].Kesti M, Müller M, Becher J, Schnabelrauch M, D’Este M, Eglin D, Zenobi-Wong M, Acta Biomater. 2015, 11, 162. [DOI] [PubMed] [Google Scholar]
- [64].Lee Y-B, Polio S, Lee W, Dai G, Menon L, Carroll RS, Yoo S-S, Exp. Neurol. 2010, 223, 645. [DOI] [PubMed] [Google Scholar]
- [65].Marco C, Joanna I, Krisztina S, Jakub J, Mariella D, Andrea B, Jan EB, Wojciech Ś, Biofabrication 2016, 8, 035002. [Google Scholar]
- [66].Zhang YS, Yue K, Aleman J, Mollazadeh-Moghaddam K, Bakht SM, Yang J, Jia W, Dell’Erba V, Assawes P, Shin SR, Dokmeci MR, Oklu R, Khademhosseini A, Ann. Biomed. Eng. 2017, 45, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Biomaterials 2016, 110, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Tamayol A, Najafabadi AH, Aliakbarian B, Arab-Tehrany E, Akbari M, Annabi N, Juncker D, Khademhosseini A, Adv. Healthc. Mater. 2015, 4, 2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Faramarzi N, Yazdi Iman K, Nabavinia M, Gemma A, Fanelli A, Caizzone A, Ptaszek Leon M, Sinha I, Khademhosseini A, Ruskin Jeremy N, Tamayol A, Adv. Healthc. Mater. 2018, 7, 1701347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Jeon O, Bouhadir KH, Mansour JM, Alsberg E, Biomaterials 2009, 30, 2724. [DOI] [PubMed] [Google Scholar]
- [71].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lim KS, Schon BS, Mekhileri NV, Brown GCJ, Chia CM, Prabakar S, Hooper GJ, Woodfield TBF, ACS Biomater. Sci. Eng. 2016, 2, 1752. [DOI] [PubMed] [Google Scholar]
- [73].Noshadi I, Hong S, Sullivan KE, Shirzaei Sani E, Portillo-Lara R, Tamayol A, Shin SR, Gao AE, Stoppel WL, Black Iii LD, Khademhosseini A, Annabi N, Biomater. Sci. 2017, 5, 2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Luiz EB, Juliana CC, Vijayan M, Ana LC, Nupura SB, Wesleyan AA, Pinar Z, Nihal EV, Amir MG, Mehmet RD, Ali K, Biofabrication 2014, 6, 024105. [Google Scholar]
- [75].Wanjun L, Zhe Z, Ning H, Yixiao Z, Lucia M, Amir KM, Alessio F, Xiangyu J, Ali K, Yu Shrike Z, Biofabrication 2018, 10, 024102. [Google Scholar]
- [76].Gao Q, He Y, Fu J.-z., Liu A, Ma L, Biomaterials 2015, 61, 203. [DOI] [PubMed] [Google Scholar]
- [77].Pati F, Jang J, Ha D-H, Won Kim S, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Nat. Commun. 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kim BS, Kim H, Gao G, Jang J, Cho DW, Biofabrication 2017, 9, 034104. [DOI] [PubMed] [Google Scholar]
- [79].Choudhury D, Tun HW, Wang T, Naing MW, Trends in Biotechnology 2018, 36. [DOI] [PubMed] [Google Scholar]
- [80].Pati F, Chow DW, Bioprinting of 3D tissue models using decellularized extracellular matrix bioink In 3D Cell Culture. Methods in Molecular Biology, K. Z, Ed. Humana Press: New York, NY, 2017; Vol. 1612, pp 381–390. [DOI] [PubMed] [Google Scholar]
- [81].Skardal A, Devarasetty M, Kang H-W, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, Atala A, Acta Biomaterialia 2015, 25. [DOI] [PubMed] [Google Scholar]
- [82].Jang J, Kim TG, Kim BS, Kim S-W, Kwon S-M, Cho D-W, Acta Biomaterialia 2016, 33. [DOI] [PubMed] [Google Scholar]
- [83].Das S, Jang J, Journal of 3D Printing in Medicine 2018, 2. [Google Scholar]
- [84].Kwee BJ, Mooney DJ, Curr. Opin. Biotechnol. 2017, 47, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Skardal A, Atala A, Ann. Biomed. Eng. 2015, 43, 730. [DOI] [PubMed] [Google Scholar]
- [86].Stanton MM, Samitier J, Sanchez S, Lab Chip 2015, 15, 3111. [DOI] [PubMed] [Google Scholar]
- [87].Merceron TK, Morgan B, Young-Joon S, Hyun-Wook K, Sang Jin L, James JY, Anthony A, Biofabrication 2015, 7, 035003. [Google Scholar]
- [88].Lombardo Vince M, Dhulst Elizabeth A, Leitsch Emily K, Wilmot N, Heath William H, Gies Anthony P, Miller Matthew D, Torkelson John M, Scheidt Karl A, Eur. J. Org. Chem. 2015, 2015, 2791. [Google Scholar]
- [89].Jeon O, Samorezov JE, Alsberg E, Acta Biomater. 2014, 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Luo Y, Luo G, Gelinsky M, Huang P, Ruan C, Mater. Lett. 2017, 189, 295. [Google Scholar]
- [91].García-Lizarribar A, Fernández-Garibay X, Velasco-Mallorquí F, Castaño AG, Samitier J, Ramon-Azcon J, Macromol. Biosci. 2018, 18, 1800167. [DOI] [PubMed] [Google Scholar]
- [92].He Y, Wu Y, Fu J.-z., Gao Q, Qiu J.-j., Electroanalysis 2016, 28, 1658. [Google Scholar]
- [93].Costantini M, Testa S, Mozetic P, Barbetta A, Fuoco C, Fornetti E, Tamiro F, Bernardini S, Jaroszewicz J, Święszkowski W, Trombetta M, Castagnoli L, Seliktar D, Garstecki P, Cesareni G, Cannata S, Rainer A, Gargioli C, Biomaterials 2017, 131, 98. [DOI] [PubMed] [Google Scholar]
- [94].Chiantore O, Guaita M, Trossarelli L, Makromol. Chem. 1979, 180, 969. [Google Scholar]
- [95].Müller M, Becher J, Schnabelrauch M, Zenobi-Wong M, J. Vis. Exp. 2013, 50632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Celikkin N, Simó Padial J, Costantini M, Hendrikse H, Cohn R, Wilson JC, Rowan EA, Święszkowski W, Polymers 2018, 10, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA, ACS Biomater. Sci. Eng. 2016, 2, 1743. [DOI] [PubMed] [Google Scholar]
- [98].Highley Christopher B, Rodell Christopher B, Burdick Jason A, Adv. Mater. 2015, 27, 5075. [DOI] [PubMed] [Google Scholar]
- [99].Popov A, Malferrari S, Kalaskar DM, J. 3D Print. Med. 2017, 1, 191. [Google Scholar]
- [100].Gao G, Cui X, Biotechnol. Lett. 2016, 38, 203. [DOI] [PubMed] [Google Scholar]
- [101].Cvetkovic C, Raman R, Chan V, Williams BJ, Tolish M, Bajaj P, Sakar MS, Asada HH, Saif MTA, Bashir R, Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Morimoto Y, Onoe H, Takeuchi S, Sci. Robot. 2018, 3, 1. [DOI] [PubMed] [Google Scholar]
- [103].Raman R, Cvetkovic C, Uzel SGM, Platt RJ, Sengupta P, Kamm RD, Bashir R, Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Raman R, Grant L, Seo Y, Cvetkovic C, Gapinske M, Palasz A, Dabbous H, Kong H, Pinera PP, Bashir R, Adv. Healthc. Mater. 2017, 6, 1700030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Miji Y, Hyeongjin L, Geun Hyung K, Biofabrication 2016, 8, 035021. [Google Scholar]
- [106].Yeo M, Kim G, ACS Biomater. Sci. Eng. 2018, 4, 728. [DOI] [PubMed] [Google Scholar]
- [107].Kim W, Kim M, Kim Geun H, Adv. Funct. Mater. 2018, 28, 1800405. [Google Scholar]
- [108].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol. 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [109].Choi Y-J, Kim Taek G, Jeong J, Yi H-G, Park Ji W, Hwang W, Cho D-W, Adv. Healthc. Mater. 2016, 5, 2636. [DOI] [PubMed] [Google Scholar]
- [110].Kim JH, Seol Y-J, Ko IK, Kang H-W, Lee YK, Yoo JJ, Atala A, Lee SJ, Sci. Rep. 2018, 8, 12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Dixon TA, Cohen E, Cairns DM, Rodriguez M, Mathews J, Jose RR, Kaplan DL, Tissue Eng. Part C Meth. 2018, 24, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhou G JH, Yin Z, Liu Y, Zhang Q, Zhang C, Pan B, Zhou J, Zhou X, Sun H, Li D, He A, Zhang Z, Zhang W, Liu W, Cao Y, EBioMedicine 2018, 28, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].M. Placko New 3D printed nose implant is fully functional. https://www.geek.com/news/new-3d-printed-nose-implant-is-fully-functional-1643508/.
- [114].Raya-Rivera AM, Esquiliano D, Fierro-Pastrana R, López-Bayghen E, Valencia P, Ordorica-Flores R, Soker S, Yoo JJ, Atala A, Lancet 2014, 384, 329. [DOI] [PubMed] [Google Scholar]
- [115].Lee VK, Dai G, Ann. Biomed. Eng. 2017, 45, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kaushik G, Leijten J, Khademhosseini A, Stem Cells 2016, 35, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sears NA, Seshadri DR, Dhavalikar PS, Cosgriff-Hernandez E, Tissue Eng. Part B Rev. 2016, 22, 298. [DOI] [PubMed] [Google Scholar]
- [118].Murphy SV, Atala A, Nat. Biotechnol. 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [119].Jang J, Yi H-G, Cho D-W, ACS Biomater. Sci. Eng. 2016, 2, 1722. [DOI] [PubMed] [Google Scholar]
- [120].Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, Li S, Belicchi M, Peretti G, Chamberlain JS, Wright WE, Torrente Y, Ferrari S, Bianco P, Cossu G, Nat. Cell Biol. 2007, 9, 255. [DOI] [PubMed] [Google Scholar]
- [121].Chal J, Al Tanoury Z, Hestin M, Gobert B, Aivio S, Hick A, Cherrier T, Nesmith AP, Parker KK, Pourquié O, Nat. Protoc. 2016, 11, 1833. [DOI] [PubMed] [Google Scholar]
- [122].Jiwlawat N LE, Jeffrey J, Van Dyke JM, Suzuki M, Stem Cells Int. 2018. 2018, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Darabi R, Arpke Robert W., Irion S, Dimos John T., Grskovic M, Kyba M, Perlingeiro Rita C. R., Cell Stem Cell 2012, 10, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Rao L TW, WeiL Y, Bao L, Chen J, Chen H, He L, Lu P, Ren J, Wu L, Luan Z, Cui C, Xiao L, Stem Cell Rev. Rep. 2012, 8, 1109. [DOI] [PubMed] [Google Scholar]
- [125].Rao L, Qian Y, Khodabukus A, Ribar T, Bursac N, Nat. Commun. 2018, 9, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ashammakhi N, Ahadian S, Zengjie F, Suthiwanich K, Lorestani F, Orive G, Ostrovidov S, Khademhosseini A, Biotechnol. J. 2018, 0, 1800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Maiullari F, Costantini M, Milan M, Pace V, Chirivì M, Maiullari S, Rainer A, Baci D, Marei HE-S, Seliktar D, Gargioli C, Bearzi C, Rizzi R, Sci. Rep. 2018, 8, 13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Hardin James O, Ober Thomas J, Valentine Alexander D, Lewis Jennifer A, Adv. Mater. 2015, 27, 3279. [DOI] [PubMed] [Google Scholar]
- [129].Ober TJ, Foresti D, Lewis JA, Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Ashammakhi N AS, Pountos I, Hu S, Tellisi N, Bandaru P, Ostrovidov S, Dokmeci M, Khademhosseini A, Biomed. Microdevices 2018. [DOI] [PubMed] [Google Scholar]
- [131].Skardal A, Mack D, Kapetanovic E, Atala A, Jackson John D, Yoo J, Soker S, Stem Cells Transl. Med. 2012, 1, 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Keriquel V, Fabien G, Isabelle A, Bertrand G, Sylvain M, Joëlle A, Jean-Christophe F, Sylvain C, Biofabrication 2010, 2, 014101. [Google Scholar]
- [133].Ram-Liebig G, Bednarz J, Stuerzebecher B, Fahlenkamp D, Barbagli G, Romano G, Balsmeyer U, Spiegeler M-E, Liebig S, Knispel H, Adv. Drug Del. Rev. 2015, 82–83, 181. [DOI] [PubMed] [Google Scholar]
- [134].Hourd P, Medcalf N, Segal J, Williams DJ, Regen. Med. 2015, 10, 863. [DOI] [PubMed] [Google Scholar]
- [135].Ladd MR, Lee SJ, Stitzel JD, Atala A, Yoo JJ, Biomaterials 2011, 32, 1549. [DOI] [PubMed] [Google Scholar]
- [136].Santhanam N, Kumanchik L, Guo X, Sommerhage F, Cai Y, Jackson M, Martin C, Saad G, McAleer CW, Wang Y, Lavado A, Long CJ, Hickman Biomaterials JJ 2018, 166, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Cvetkovic C, Rich MH, Raman R, Kong H, Bashir R, Microsyst. Nanoeng. 2017, 3, 17015. [DOI] [PMC free article] [PubMed] [Google Scholar]


