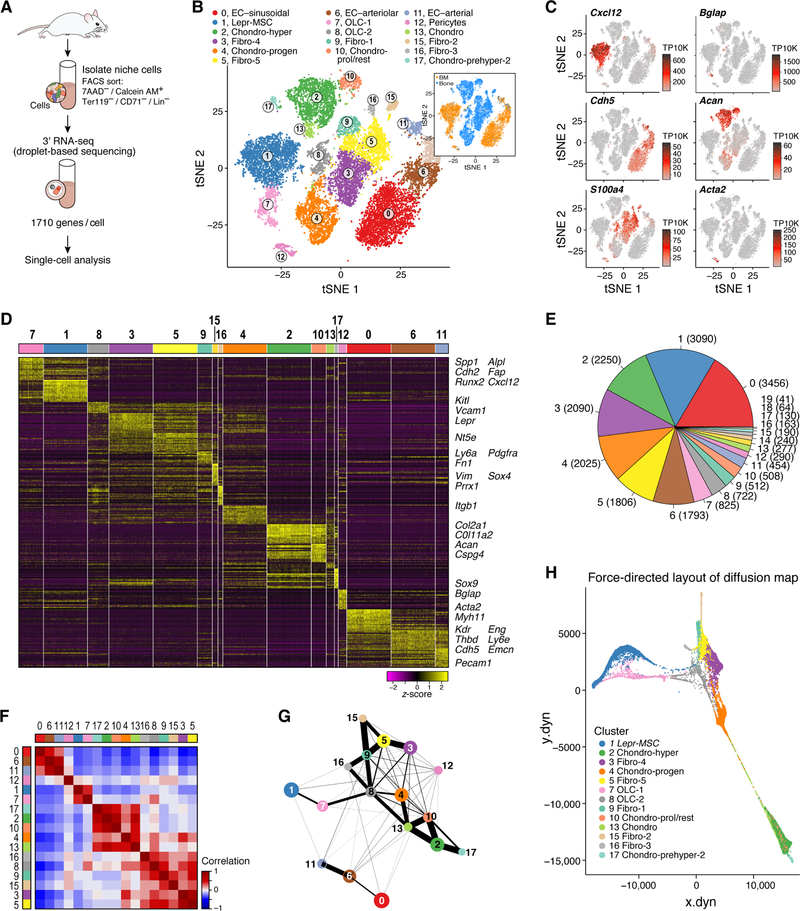
Figure 1. A single cell atlas of the mouse bone marrow stroma.
(A) Study Overview. (B,C) Seventeen bone marrow stroma cell clusters. t-SNE of 20,896 nonhematopoietic cells (mixed bone and bone marrow fractions, n=6 mice), annotated post hoc and colored by clustering or bone (~10,000, non-hematopoietic cells, n=4 mice) or bone marrow (~10,000 non-hematopoietic cells, n=4 mice) location (insert) (B) or by expression (color bar, TP10K) of key cell type marker genes (C). (D) Cluster signature genes. Expression (row-wide z-score of ln(TP10K+1)) of top differentially expressed genes (rows) across the cells (columns) in each cluster (color bar, top, as in B). The largest clusters were down-sampled. Key genes highlighted on right. (E) Number of cells in each subset. Color bar as in B. (F–G) Differentiation relations between cells or clusters. (F) Pearson’s correlation (color bar) between the average gene expression profiles of clusters (rows, columns, color code as in B). (G) Relatedness (edges; width indicates strength) between clusters (nodes, colored as in B) based on cluster graph abstraction. (H) A force directed layout embedding (FLE) of the cells (dots) from a diffusion map (50 components) computed with the cells from strongly connected clusters (as indicated in G and F, without clusters 1, 6, 11, 12).