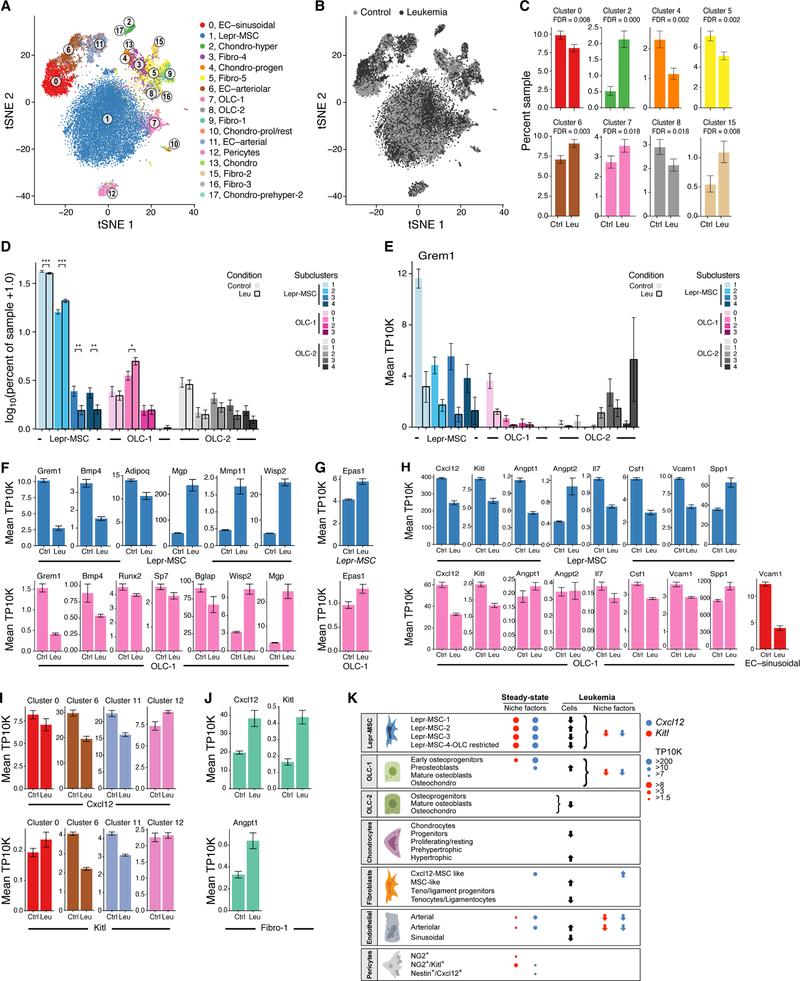
Figure 7. Remodeling of the bone marrow stroma in leukemia.
(A,B) A census of the leukemic bone marrow stroma. tSNE of 23,004 cells (dots) from mice transplanted with control (n=5, 12,456 cells) or leukemia allele bearing (n=4, 10,548 cells) bone marrow colored by cluster assignment (as in Figure 1B), or by condition (control: light grey; leukemia: dark grey). (C,D) Compositional changes in bone marrow stroma. Binomial fit mean as percent of cells (y axis) assigned to specific cluster (C) or to Lepr-MSC and OLCs subclusters (D) among control or leukemic samples (x axis). Error bars: 95% confidence interval of the binomial fit mean. (E) Changes in Grem1 expression in Lepr-MSCs and OLCs. Average of samples (TP10K, y axis) in Lepr-MSC and OLC subclusters (x axis). Error bars: SEM. (F–H) Changes in niche remodeling, hypoxia and hematopoietic regulator genes in Lepr-MSCs, OLC-1s and sBMECs in leukemia. Average expression (TP10K, y axis) of niche genes in Lepr-MSCs (top) and OLC-1s (bottom). Error bars: SEM. (I,J) Changes in Cxcl12, Kitl and Angpt1 expression in BMECs, pericytes or fibroblasts. Average expression (TP10K, y axis) of the denoted genes in specific cell clusters. Error bars: SEM. (K) A census of the bone marrow stroma in homeostasis and leukemia. Horizontal boxes: broad cell types, with subsets noted. Circles: expression levels of key niche factors, Kitl and Cxcl12. Dark arrows: changes in subset proportion in Leukemia. Colored arrows: changes in relative expression in leukemia.