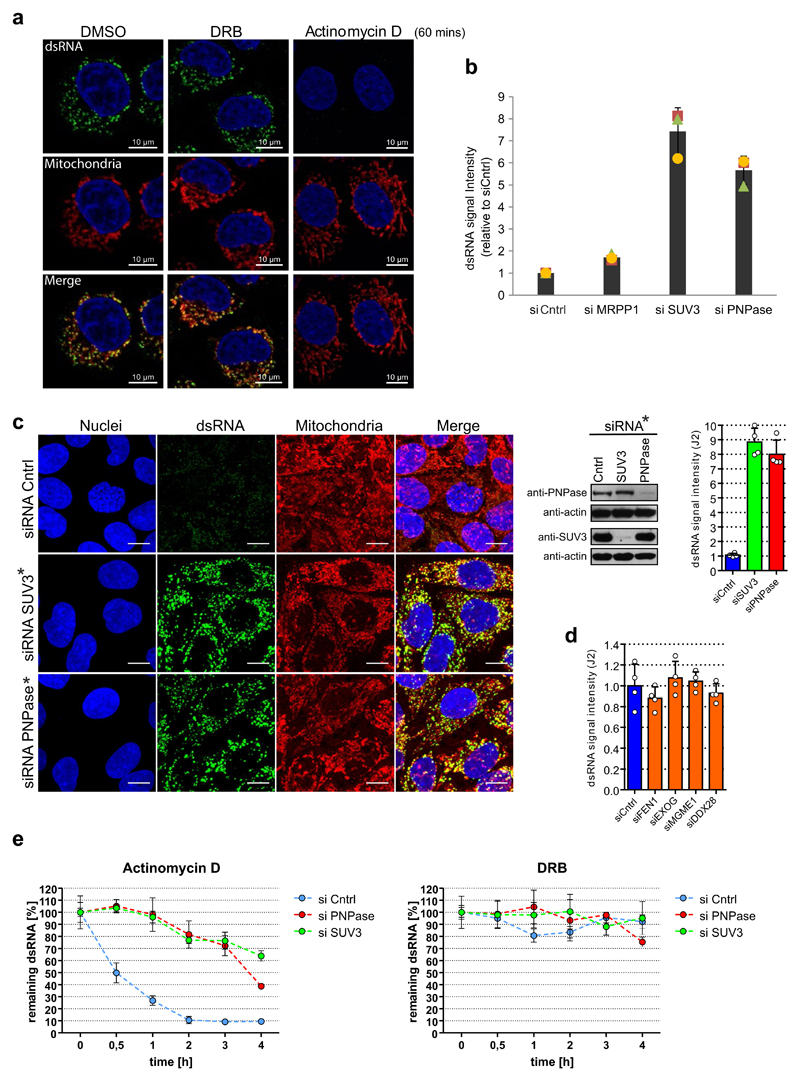
Extended Data Fig. 2. RNA degradosome components SUV3 and PNPase are involved in mtdsRNA turnover.
a, HeLa cells treated with DMSO, DRB (100 μM) and actinomycin-D (0.5 μg ml−1) for 60 min and stained with anti-dsRNA (J2) antibody (green). Mitochondria were stained with MitoTracker Red CMXRos and nuclei with DAPI (blue) (representative of two experiments). b, Flow cytometric analysis of dsRNA levels in HeLa cells treated with the indicated siRNAs. Cells were labelled with J2 antibody or an isotype control. Data are mean ± s.d. from three independent experiments. c, Left, detection of dsRNA with J2 antibody in HeLa cells after depletion of PNPase or SUV3 by On-TARGETplus siRNAs (indicated with an asterisk and listed in Extended Data Table 2). Mitochondria were stained with MitoTracker Deep Red. Nuclei are stained with Hoechst (blue). Scale bars, 10 μm. Right top, western blot showing PNPase or SUV3 depletion. Blots are representative of four experiments. For gel source images, see Supplementary Fig. 1. Far right top, Quantification of dsRNA levels in HeLa cells depleted of PNPase or SUV3. Data are mean ± s.d. from four independent experiments. d, Quantitative analysis of fluorescent signals from dsRNA in HeLa cells with depleted enzymes involved in mitochondrial nucleic acids metabolism. Data are mean ± s.d. from four independent experiments. e, HeLa cells were transfected with siRNA specific for PNPase, SUV3, or non-targeting control. Prior to fixation, cells were treated for indicated times with inhibitors of transcription: actinomycin-D (0.5 μg ml−1) or DRB (100 μM). Immunostaining of dsRNA was performed and cells were imaged using a fluorescent microscope screening station. Data are mean ± s.d. from four independent experiments.