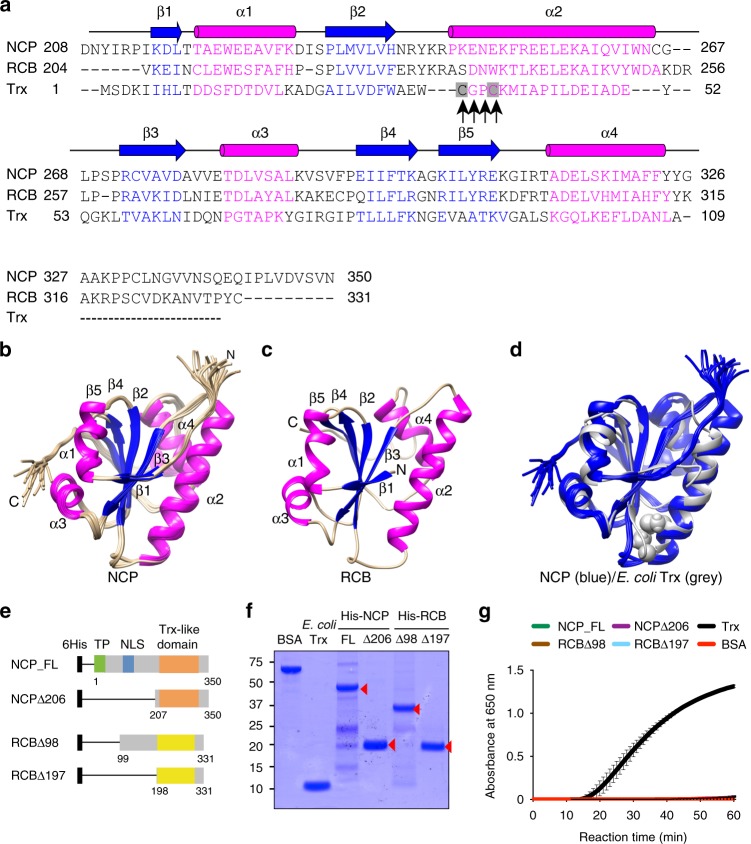
Fig. 6.
NCP and RCB structurally resemble E. coli Trx but lack reductase activity. a Sequence alignment of the Trx-like domains of NCP and RCB with that of E. coli Trx. Magenta characters represent alpha helices and blue characters represent beta sheets. The black arrows indicate the conserved catalytic -Cys-X-X-Cys- motif in E. coli Trx. b NMR structures of NCP’s Trx-like domain (PDB ID: 6NE8). c Simulated structure of RCB’s Trx-like domain based on the NMR structure of NCP. d Overlay of the structure of NCP’s Trx-like domain with the crystal structure of E. coli Trx (PDB ID: 1XOB). The two cysteines in E. coli Trx are shown. e Schematic illustration of NCP and RCB fragments used for the in vitro Trx reductase assays. f SDS-PAGE gels showing BSA (negative control), E. coli Trx (positive control), and purified His-tagged recombinant NCP and RCB fragments. g Trx activity assays showing that recombinant NCP and RCB proteins lack Trx activity. Trx activity was determined based on the increase in turbidity after the reduction of insulin, as measured by the absorbance at 650 nm53. E. coli Trx and BSA proteins were used as a positive control and a negative control, respectively. The source data of the Trx activity assay in g are provided in the Source Data file