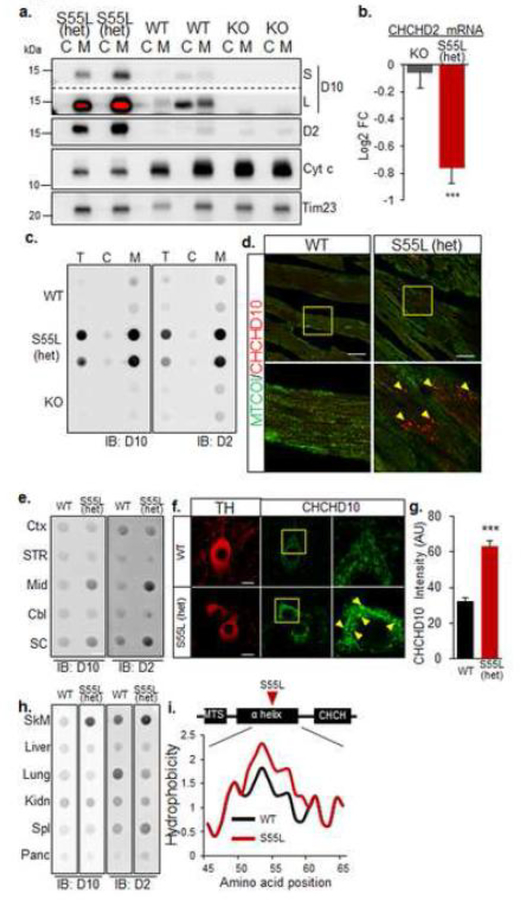
Figure 7. Mutant CHCHD10 aggregates with CHCHD2 in mitochondria of affected tissues.
A. Western blot of heart cytosolic (C) and mitochondrial (M) fractions. D10, CHCHD10; D2, CHCHD2; Cyt c, cytochrome c, short (S) and long (L) exposures are shown for D10. The red color in L indicates overexposure. The inner membrane protein Tim23 was used as a loading control. B. CHCHD2 mRNA content measured by RNA-seq and expressed as average log2 fold change relative to WT control. Error bars indicate SEM. ***p<0.001 by Student’s t-test with Benjamini-Hochberg correction. C. Filter trap analysis of total homogenate (T), cytosolic (C) and mitochondrial (M) fractions from heart. Immunoreactive material was detected by immunoblot (IB) with either D10 or D2 antibodies. D. Cardiomyocytes immunolabeled with antibodies against CHCHD10 (red) and cytochrome oxidase subunit 1 (MTCOI). Bars = 25 μm. The lower panel in each image is a magnification of the area indicated by the square in the top panel. E. Filter trap analysis of specific brain regions for D10 and D2 aggregates. Immunoreactive material was detected by immunoblot (IB) with either D10 or D2 antibodies. Mid, midbrain; SC, spinal cord; CTX, cortex; Str, striatum, Cbl, cerebellum. F. Substantia nigra neurons immunostained for TH and CHCHD10. Bars = 10 μm. The right panel in each image is a magnification of the area indicated by the square in the middle panel. Hyper-intense CHCHD10 positive puncta are indicated by arrows. G. Average CHCHD10 immunolabeling intensity in TH+ neurons of the substantia nigra. n=20 WT and n=35 CHCHD10S55L TH+ neurons, from 4 animals per group. Error bars indicate SEM. ***p<0.001 by Student’s t-test. H. Filter trap analysis of peripheral tissues for D10 and D2. SkM, skeletal muscle; Kdn, kidney; Spl, spleen Panc, pancreas. I. Kyte-Doolittle hydrophobicity plots of the α-helices of CHCHD10 WT and CHCHD10S55L.