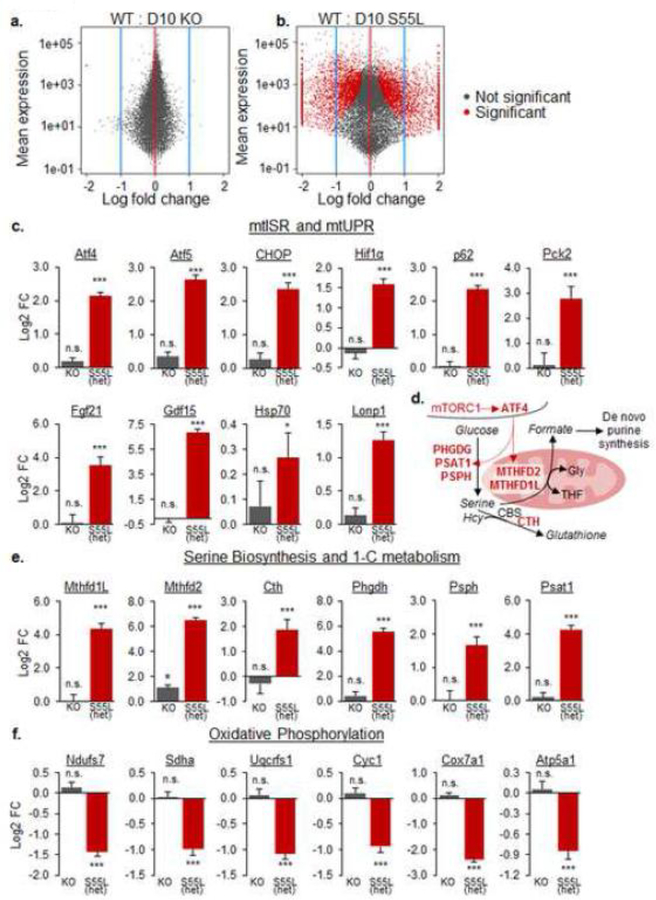
Figure 9. CHCHD10S55L hearts activate a transcriptional profile of mtISR.
A. and B. Volcano plots of transcriptomic changes in CHCHD10 KO and CHCHD10S55L hearts relative to WT heart. C. Changes (log2 fold) in heart RNA content of genes involved in mtUPR and mtISR. Hif1α, hypoxia-induced factor 1α; Pck2, Phosphoenolpyruvate Carboxykinase 2; Hsp70, heat shock protein 70; Lonp1, Lon protease homolog. n.s. not significant. *p<0.05, ***p<0.001, by Student’s t-test with Benjamini-Hochberg correction. D. Schematic diagram of enzymes involved in serine biosynthesis and one-carbon metabolism under ATF4 transcriptional regulation. PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; MTHFD1L, methylenetetrahydrofolate dehydrogenase 1-like; CTH, Cystathionine gamma-lyase. E. Changes (log2 fold) in heart RNA content of genes involved in serine biosynthesis and one-carbon metabolism. ***p<0.001, by Student’s t-test with Benjamini-Hochberg correction. F. Changes (log2 fold) in heart RNA content of genes encoding subunits of the mitochondrial oxidative phosphorylation complex I (Ndufs7), II (Sdha), III (Uqcrfs1), IV (Cox7a1), and V (Atp5a1) and cytochrome c (cyc1). ***p<0.001, by Student’s t-test with Benjamini-Hochberg correction.