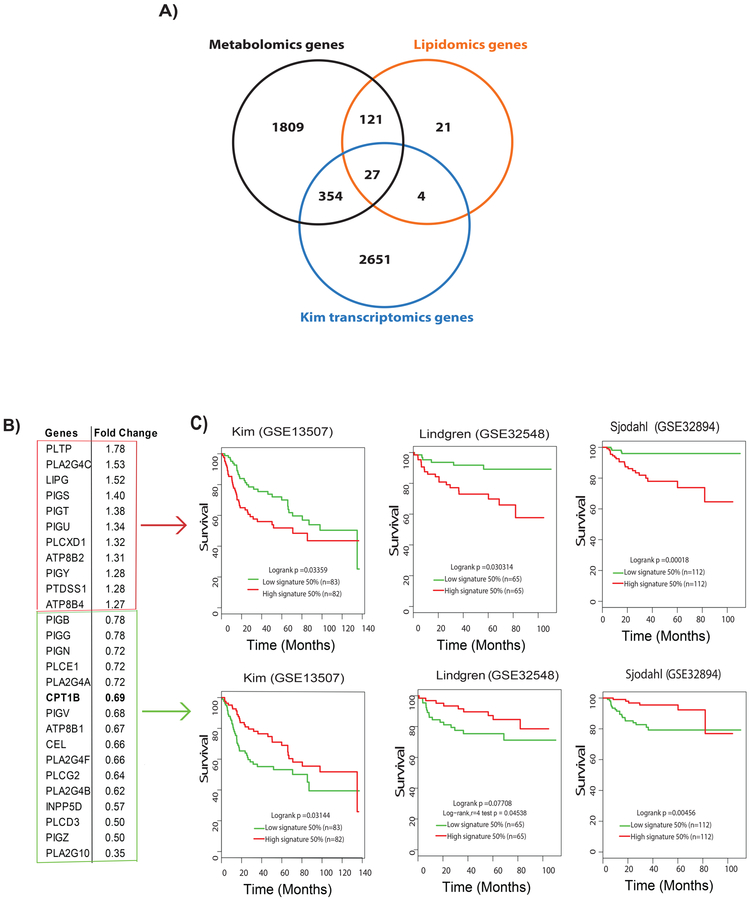
Figure 3. Metabolomic-lipidomic–transcriptomic integration strategies to identify potential prognostic gene signature for BLCA.
A) The integration of a significant genes derived from metabolomics, lipidomics, and transcriptomics data comparing low-grade and high-grade BLCA resulted in a set of 27 common genes. B) Eleven upregulated and 16 down-regulated genes in high-grade BLCA in the Kim cohert (GSE13507) with their fold change. C) Survival analysis shows poor survival associated with the upregulated gene signature (Kim, log rank p =0.03359;, Lindgren (GSE32584), log-rank p=0.030314;, Sjodahl (GSE32894), log-rank p=0.00018) and downregulated gene signature (Kim, log-rank p=0.03144;, Lindgren, log-rank p=0.07708; and Sjodahl, log-rank p=0.00456.