Figure 1. Residue level binary classification of order/disorder.
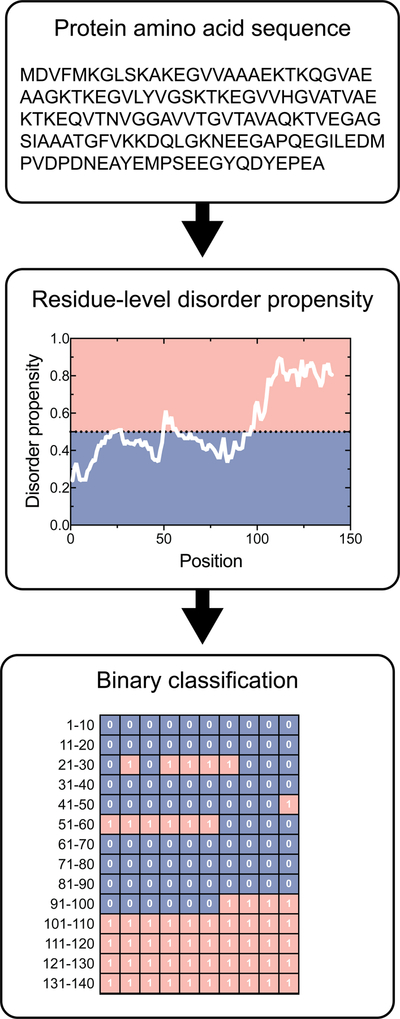
The amino acid sequence of a protein of interest is input into a disorder prediction algorithm, which returns residue-by-residue disorder propensity values. These disorder propensity values are compared against a defined threshold to classify a residue as either ordered or disordered. IUPred-disorder predictions are displayed for the H. sapiens alpha-synuclein protein (P37840). IUPred uses a threshold value of 0.5 to assign ordered and disordered residues. Residues that fall within the red region are disordered, whereas residues that fall in the blue region are ordered.
