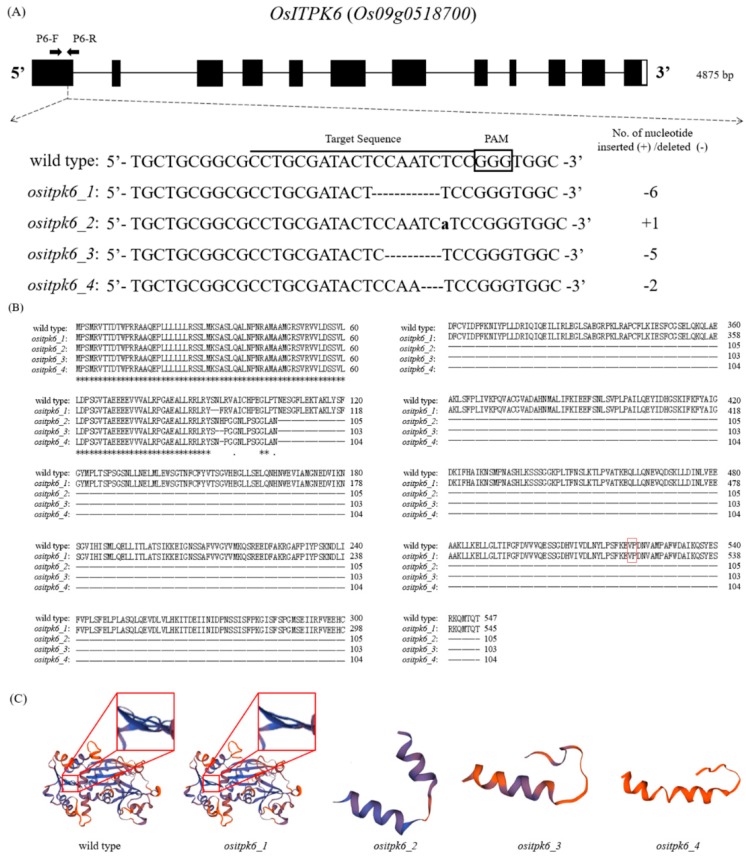
Figure 1.
Schematic diagram of OsITPK6 and sgRNA target site for CRISPR/Cas9-mediated mutagenesis of OsITPK6, and prediction of the related wild-type and mutant proteins. (A) Exons, introns, and UTRs are indicated by solid boxes, lines, and blank boxes, respectively. P6-F and P6-R are primers for mutation genotyping, and their positions are indicated by arrowheads. Mutation identified within the target site of OsITPK6 generated through CRISPR/Cas9-mediated genome editing in rice. The PAM sequences (NGG) are boxed, and the 20-nt target sequences are underlined. Mutations are shown in lowercase letters (for insertions) or ‘–’ (for deletions). (B) The amino acid sequences of mutant proteins were aligned to that of the wild-type protein using the Clustal Omega Multiple Sequence Alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/). The numbers represent the total number of amino acids, and the amino acid V521 and P522 are highlighted in red box. (C) The three-dimensional structures of OsITPK6 and its mutants were analyzed on SWISS-MODEL (https://www.swissmodel.expasy.org/).