Abstract
NMR has provided a wealth of structural and dynamical information for RNA molecules of up to ~50 nucleotides, but its application to larger RNAs has been hampered in part by difficulties establishing global structural features. A potential solution involves measurement of NMR perturbations after site-specific paramagnetic labeling. Although the approach works well for proteins, the inability to place the label at specific sites has prevented its application to larger RNAs transcribed in vitro. Here, we present a strategy in which RNA loop residues are modified to promote binding to a paramagnetically tagged reporter protein. Lanthanide-induced pseudocontact shifts are demonstrated for a 232-nucleotide RNA bound to tagged derivatives of the spliceosomal U1A RNA binding domain. Further, the method is validated a 36-nucleotide RNA for which measured NMR values agreed with predictions based on the previously known protein and RNA structures. The ability to readily insert U1A binding sites into ubiquitous hairpin and/or loop structures should make this approach broadly applicable for the atomic-level study of large RNAs.
Graphical Abstract
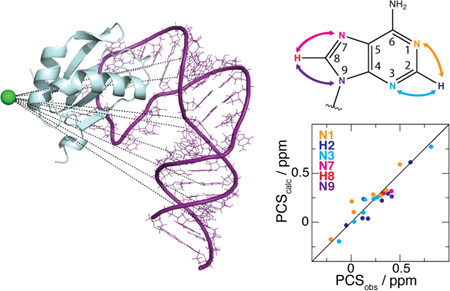
RNA plays a crucial role in many cellular processes, including translation, transcription, enzymatic catalysis, and genetic regulation.1 Knowledge of RNA structure and dynamics is important not only for understanding of biological mechanisms but also for the development of novel RNA-based therapeutics. These efforts are hampered by difficulties applying current biophysical techniques to this class of molecule. For example, RNA flexibility can inhibit crystallization needed for X-ray structure determination, while intrinsic low proton density and poor chemical shift dispersion create challenges for solution-state nuclear magnetic resonance (NMR) spectroscopy.2 A particularly serious hurdle for larger RNAs (> ~100 nt) is the typical lack of experimental information defining the relative configuration of secondary structure elements. Although local structural features can be well defined for larger RNAs using 2H-edited NMR approaches,3–6 segmental labeling,7 or site-specific labeling,8 interproton distances across neighboring helices are typically too large for measurement by nuclear Overhauser effects (NOEs), and until recently,9 rapid 1H–13C NMR relaxation and other technical problems have limited collection of residual dipolar couplings (RDCs) to relatively small RNAs. These limitations have led to the use of hybrid methods, in which high-resolution local structural information is complemented with lower-resolution global information derived from small angle X-ray scattering (SAXS) or cryo-electron microscopy.10–14 New NMR methods for capturing elusive long-range structural information would clearly be beneficial.
The large magnetic moment of an unpaired electron attached to a molecule of interest has profound effects on the associated (paramagnetic) NMR spectra. Such effects are long-reaching and may manifest themselves as spectral line broadening (e.g., paramagnetic relaxation enhancements, PREs), line shifts (e.g., pseudocontact shifts, PCSs), and partial molecular alignment under the external magnetic field (from which, e.g., RDCs can be obtained). The known structure dependence of these NMR observables thus offers a wealth of long-range information. PREs and PCSs can provide structural information for distances up to 35 Å15 and 56 Å16, respectively, while RDCs report on the whole molecule. Paramagnetic NMR has emerged as a powerful tool to study proteins17–18 thanks, in part, to the continued development of paramagnetic tags that efficiently and robustly attach in a covalent fashion to engineered surface-exposed cysteines of otherwise diamagnetic molecules. By contrast, paramagnetic applications to RNA suffer from issues associated with this direct chemical tagging, including sample instability and low tagging efficiency. Although techniques are available to incorporate modified bases in a site-specific manner,19–21 these often involve relatively low-yielding multi-step enzymatic or synthetic protocols. Perhaps the biggest challenge is the inability to tag specific sites in large RNAs prepared by in vitro transcription. For example, although there are dozens of lanthanide-based tags for measuring PCSs in proteins, the development of a nucleic acid counterpart is still in an embryonic state.22
Here, we present a general strategy to induce paramagnetic effects on RNA that circumvents the above-mentioned pitfalls of direct tagging. It comprises replacement of a segment of the RNA of interest by another that contains the specific binding sequence for a small protein domain. The protein is independently paramagnetically tagged in a conventional manner and, upon complexation with the chimeric RNA, indirectly provides the sought-after long-range structural information on the target RNA via intermolecular effects. The protein chosen for this purpose is the RNA-binding domain of the U1A spliceosomal protein (henceforth “U1A”) for several reasons: (i) it specifically and tightly binds its cognate RNA (KD ~ 10–11 M),23 (ii) it has been previously structurally characterized both in isolation24 and in complex with a 21-nucleotide (nt) hairpin bearing the binding sequence25 (Figure 1A, B), (iii) it is relatively small (11 kDa), thus limiting spectral deterioration and perturbation of the target RNA structure, and (iv) it is straightforward to express and purify due to its high solubility.26 In addition, the U1A binding loop has previously been successfully engineered into a number of RNA molecules, allowing complexation with U1A as an aid to crystallization.27
Figure 1.
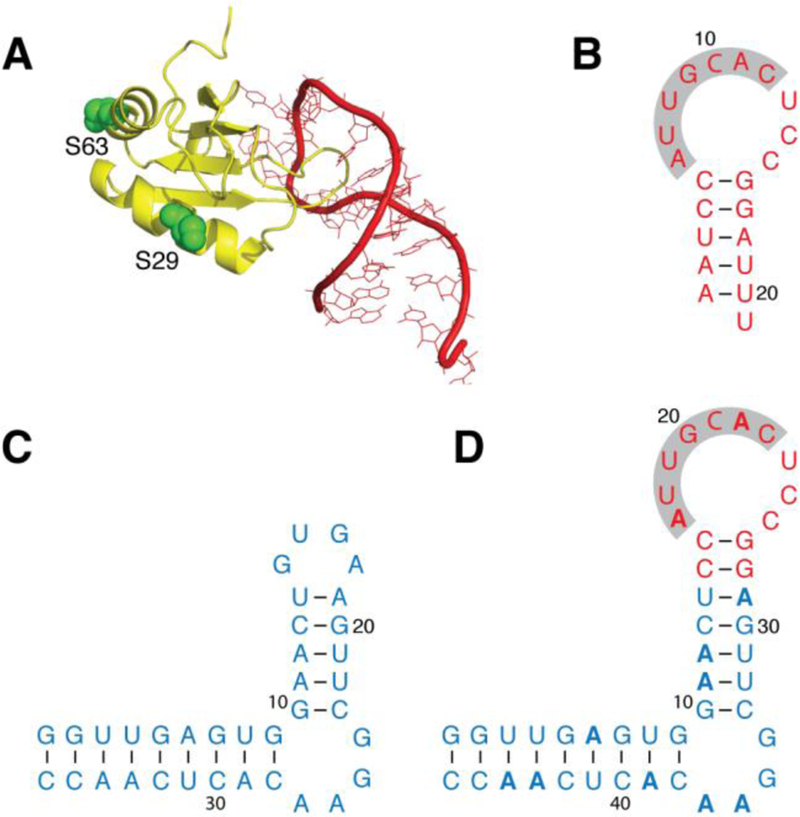
The binding of U1A to a specific RNA sequence is used to paramagnetically study a chimera that contains the RNA sequence of interest. (A) X-ray structure (PDB ID: 1URN) of U1A (yellow; chain B) bound to a 21-nt RNA hairpin (red; chain Q). U1A side chains of S29 and S63 (green) indicate the positions used for paramagnetic tagging. (B) Construct of the RNA hairpin shown in panel A; the seven nucleotides that form close contacts with U1A are highlighted. (C) SLCA RNA construct. (D) Chimeric RNA generated from the SLCA (blue) and hairpin (red) sequences, with 15N isotopic enrichment of adenosines only (indicated in bold). All RNA constructs (panels B–D) indicate base pairing (dashes).
Two alternative paramagnetic tagging sites on U1A were explored, involving mutants S29C and S63C (see Figure 1A). The tag chosen for the present study is the recently introduced M8-DOTA-SPy,28–29 which chelates paramagnetic lanthanide ions within a rigid lanthanide cage that discourages conformational exchange. M8-DOTA-SPy has been recently used to characterize intrinsically disordered proteins30 and dynamic protein–protein complexes.31 PCSs were measured by taking chemical shift differences between samples incorporating paramagnetic (Tm3+-loaded) and diamagnetic (Lu3+-loaded) versions of the tag. For a given nucleus, the PCS depends on the length, r, and orientation of the lanthanide–nucleus vector relative to the principal axes of the (paramagnetic) lanthanide’s magnetic susceptibility tensor via:32
| (1) |
where (r, θ, ϕ) are the spherical coordinates of the vector, and and are the axial and rhombic components of the tensor, respectively.
We tested the feasibility of this approach for large RNAs using a 232-nt, 78 kDa HIV-1 Rev response element RNA construct engineered to adopt one of two equilibrium conformations (RRE232B),33 in which the stem IIb loop was replaced by the U1A binding loop. NMR characterization of the resulting 241-nt chimera (SI Figure S1) was based on a sample 15N-enriched at adenosine positions only, for which sensitive detection of slowly relaxing H2 signals via the two-bond H2 to N1/N3 scalar couplings is feasible in large RNAs.9,34 Figure 2A shows that PCSs are readily detected in the 1H/15N SOFAST-HMQC34–35 spectrum. It is not possible to validate these measurements directly as a structure is not yet available for this construct, so our analysis focused on a 36-nt RNA based on stem-loop C from the Moloney Murine Leukemia Virus 5’-Leader (SLCA; Figure 1C). The structure of SLCA has previously been elucidated by NMR with long-range information from RDCs9 and therefore serves as a suitable validation system for this approach. For this construct the distal loop of SLCA (see Figure 1C) is replaced by the binding loop of the U1A ligand (Figure 1B), yielding a 46-nt chimera (Figure 1D).
Figure 2.
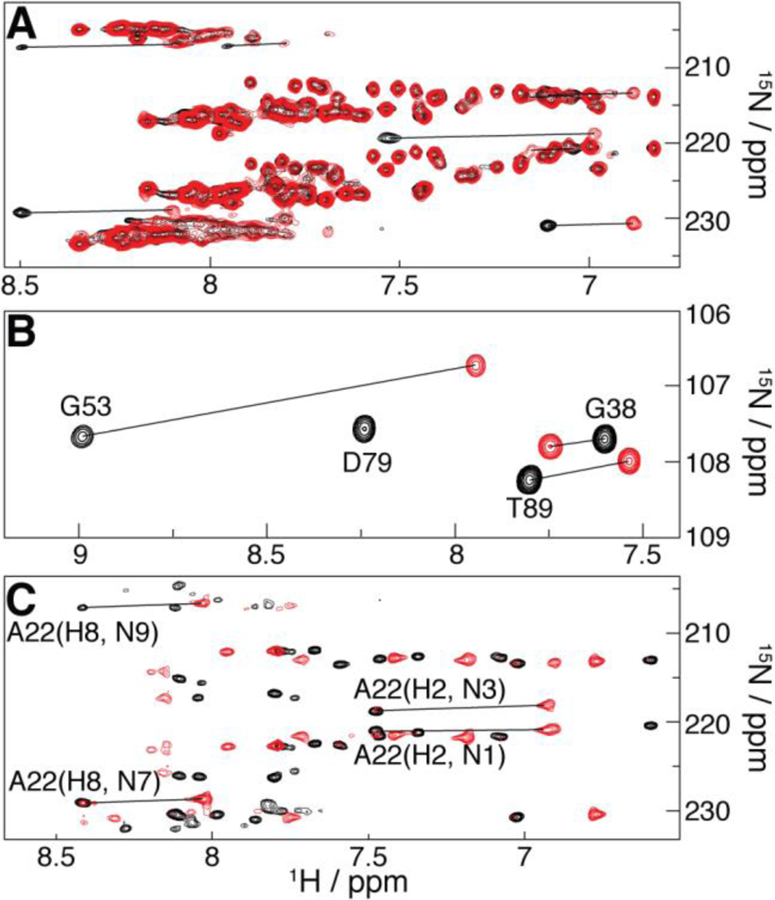
PCSs evidenced by superposition of diamagnetic (Lu3+-based; black) and paramagnetic (Tm3+-based; red) spectra involving U1A S29C tagged with M8-DOTA-SPy. (A) Region of the 1H/15N SOFAST-HMQC spectra of 15N-adenine RRE232B-based RNA chimera bound to natural abundance U1A. (B) Region of the 1H/15N-HSQC spectra of 15N-U1A bound to the unlabeled 21-nt hairpin (intramolecular PCSs). (C) Region of the 2J 1H/15N-HMQC spectra of the 15N-adenine SLCA-based RNA chimera (intermolecular PCSs) bound to natural abundance U1A. Selected peak assignments and PCSs (lines) are indicated.
Figure 2B shows PCSs in the 1H/15N-HSQC spectrum of uniformly 15N-enriched U1A S29C bound to its cognate RNA (Figure 1B) at natural isotopic abundance. Such intramolecular PCSs are observed throughout the U1A S29C HSQC spectrum, as well as that of the S63C mutant (SI Figure S2). Intermolecular PCSs for the 15N-adenosine labeled SLCA-based chimeric RNA (Figure 1D) were measured upon complexation with natural-isotopic-abundance U1A S29C (Figure 2C) and U1A S63C (SI Figure S3), using a 2J 1H/15N-HMQC experiment, yielding correlations between adenine’s H2–N1/N3 and H8–N7/N9, and thus potentially six PCSs per base (see SI Tables S1–4 for assignments).
The PCSs from each U1A tagging position underwent fitting to a structural model of the complex between the RNA chimera and U1A. The RNA was modeled using established protocols,36 taking the constituent SLCA and U1A-binding hairpin sequences to adopt the conformations observed in their native constructs (Figure 3A and 1A, respectively), and the adjoined SLCA and hairpin stems to form an uninterrupted A-form helix.
Figure 3.
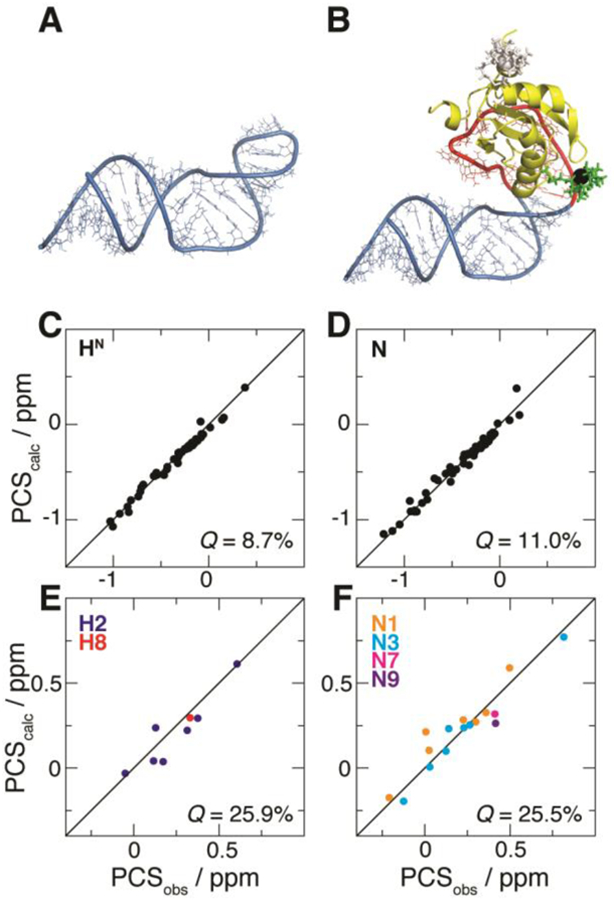
Fit of PCSs from U1A S29C against the structural model of the complex between U1A and the SLCA-based RNA chimera. (A) NMR structure of SLCA.9 (B) Modeled complex between U1A (yellow) and the RNA chimera built with atomic configurations from SLCA (blue) and the 21-nt hairpin (red). The optimized M8-DOTA-SPy conformation is shown (lanthanide, black sphere; remainder, green sticks). For comparison, the conformation of the tag at U1A position 63 (optimized with its corresponding PCSs) is shown (white). (C–F) Correlation plots between observed (PCSobs) and calculated (PCScalc) PCSs for (C) 1H and (D) 15N atoms of U1A (yellow in (B), respectively), and for (E) 1H and (F) 15N atoms of SLCA RNA (blue in (B)). Associated Q-factors are indicated.
Consequently, the model preserves the interhelical configuration of the reference SLCA RNA structure. U1A was positioned by reference to the high-resolution structure in complex with its cognate RNA25 (Figure 1A). The structural model (shown in Figure 3B) served as the basis for the PCS fit, achieved by optimizing the conformation of the M8-DOTA-SPy tag against the corresponding PCS data, along with the direction, and the axial and rhombic components of the susceptibility tensor (see Equation 1), as previously described37 (see also Supporting Information).
For each tagging site, simultaneous fitting of all the associated PCS datasets was attempted. In the case of the U1A S63C mutant, however, inclusion of the RNA 15N dataset yielded poor results overall (not shown), due to non-negligible contributions from residual chemical shift anisotropy (RCSA)38 to this weak intermolecular PCS dataset. By contrast, this behavior was not observed for intramolecular data (from both tag positions) or intermolecular data from U1A S29C, where 15N PCSs are substantially larger than the RCSA effects because of the closer proximity of the tag (see Figure 3B). Thus, the fitting procedure based on U1A S63C PCSs subsequently excluded the above-mentioned offending dataset, which will not be considered any further. Q-factors39 are used to indicate the quality of agreement between observed PCSs and those fit to the structural model of the complex. For example, PCSs stemming from U1A S29C exhibit Q-factors of 25.9% and 25.5% (for 1H and 15N, respectively) for the SLCA RNA data, and 8.7% (1H) and 11.0% (15N) for the U1A protein (see SI Table S5 and Figure 3C–F). Similar observations apply to the U1A S63C-based PCS dataset (SI Table S5 and SI Figure S4).
Here, we have presented a strategy to obtain long-range RNA structural information via paramagnetic effects introduced by the reporter protein U1A, which thus bypasses the common hurdles of direct RNA chemical tagging. Although, to our knowledge, never applied to nucleic acids, similar methodology has been previously implemented in protein NMR research.40–41 Further, the addition of extraneous RNA to modify diamagnetic alignment to facilitate its structural and dynamical characterization by NMR has been previously documented.42–43
In the current study, indirect PCSs induced by the M8-DOTA-SPy tag28 are shown for a 232-nt RNA derived from the HIV-1 RRE, demonstrating the feasibility of our approach for large RNAs. When attached at two alternative U1A locations, measurements agree with a model based on the known, reference RNA structure of SLCA,9 validating the use of this method for generating long-range structural information. Since the U1A tagging sites were chosen so that the tag does not sterically contact the RNA, the tag conformation may be independently determined with intramolecular (U1A) data only, using a generic RNA binding partner such as the 21-nt hairpin employed here (see SI Figure S5). Therefore, an arbitrary structure determination scenario based on M8-DOTA-SPy attached at these U1A locations may constrain the tag conformations to those precomputed with the intramolecular PCSs presented here, reducing both data requirements (as only the intermolecular, RNA-specific PCSs are needed) and the number of fit parameters in the structure calculation.44 Indeed, a reliable simultaneous optimization of the susceptibility tensor parameters, tag and RNA conformations against RNA-only PCSs may not be possible due to the general low density of NMR observables, available for large RNAs—an issue that would persist in the event of a successful direct RNA tagging procedure. Although demonstrated here with M8-DOTA-SPy, our method is general, and makes the arsenal of protein-specific paramagnetic tags17–18 accessible for RNA research, offering a new avenue for the structural and dynamical characterization of large RNAs.
Supplementary Material
ACKNOWLEDGMENT
We thank the following people from the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health (NIH): Dr. Duck-Yeon Lee of the Biochemistry Core Facility for expertise regarding LC−MS, Dr. Adrian Ferré-D’Amaré of the Biochemistry and Biophysics Center for the U1A plasmid and helpful discussion, and Dr. Ana C. L. Opina, Dr. Olga Vasalatiy, and Dr. Rolf Swenson of the Imaging Probe Development Center for synthesizing the lanthanide tags.
Funding Sources
This work was supported by the Intramural research program of the National Heart, Lung, and Blood Institute to N.T., CIT, NCI, NHLBI, and NIDDK to G.A.B., by the NIH grants U54 GM103297 to JM and GM42561 to MFS.
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
Materials and Methods, Figures with NMR and structural calculation data, Tables of chemical shift assignments for RNA and Q-factors for PCS data (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Gesteland RF; Cech TR; Atkins JF, THE RNA WORLD THIRD EDITION The Nature of Modern RNA Suggests a Prebiotic RNA World Preface. Cold Spring Harb Mon 2006, 43, Ix–X. [Google Scholar]
- 2.Barnwal RP; Yang F; Varani G, Applications of NMR to structure determination of RNAs large and small. Arch Biochem Biophys 2017, 628, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Souza V; Dey A; Habib D; Summers MF, NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol 2004, 337 (2), 427–42. [DOI] [PubMed] [Google Scholar]
- 4.Keane SC; Heng X; Lu K; Kharytonchyk S; Ramakrishnan V; Carter G; Barton S; Hosic A; Florwick A; Santos J; Bolden NC; McCowin S; Case DA; Johnson BA; Salemi M; Telesnitsky A; Summers MF, Structure of the HIV-1 RNA packaging signal. Science 2015, 348 (6237), 917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu K; Heng X; Garyu L; Monti S; Garcia E; Kharytonchyk S; Dorjsuren B; Kulandaivel G; Jones S; Hiremath A; Sachin Divakaruni S; LaCotti C; Barton S; Tummillo D; Hosic A; Edme K; Albrecht S; Telesnitsky A; Summers MF, NMR detection of structures in the HIV-1 5´-leader RNA that regulate genome packaging. Science 2011, 344, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu K; Heng X; Summers MF, Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 2011, 410, 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duss O; Maris C; von Schroetter C; Allain FH, A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res 2010, 38 (20), e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y; Holmstrom E; Zhang J; Yu P; Wang J; Dyba MA; Chen D; Ying J; Lockett S; Nesbitt DJ; Ferre-D’Amare AR; Sousa R; Stagno JR; Wang YX, Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 2015, 522 (7556), 368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchant J; Bax A; Summers MF, Accurate Measurement of Residual Dipolar Couplings in Large RNAs by Variable Flip Angle NMR. J Am Chem Soc 2018, 140 (22), 6978–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki Y; Irobalieva RN; Tolbert BS; Smalls-Manty A; Iyalla K; Loeliger K; D’Souza V; Khant H; Schmid MF; Garcia E; Telesnitsky A; Chiu W; Summers MF, Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol 2010, 404, 751–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K; Keane SC; Su Z; Irobalieva RN; Chen M; Van V; Sciandra CA; Marchant J; Heng X; Schmid MF; Case DA; Ludtke SJ; Summers MF; Chiu W, Structure of the 30 kDa HIV-1 RNA Dimerization Signal by a Hybrid Cryo-EM, NMR, and Molecular Dynamics Approach. Structure 2018, 26 (3), 490–498 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J; Zuo X; Yu P; Xu H; Starich MR; Tiede DM; Shapiro BA; Schwieters CD; Wang YX, A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J Mol Biol 2009, 393 (3), 717–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YX; Zuo X; Wang J; Yu P; Butcher SE, Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods 2010, 52 (2), 180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo X; Wang J; Foster TR; Schwieters CD; Tiede DM; Butcher SE; Wang Y-X, Global molecular structure and interfaces: Refining an RNA:RNA complex structure using solution X-ray scattering data. J Am Chem Soc 2008, 130, 3292–3293. [DOI] [PubMed] [Google Scholar]
- 15.Clore GM; Iwahara J, Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 2009, 109 (9), 4108–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keizers PH; Mersinli B; Reinle W; Donauer J; Hiruma Y; Hannemann F; Overhand M; Bernhardt R; Ubbink M, A solution model of the complex formed by adrenodoxin and adrenodoxin reductase determined by paramagnetic NMR spectroscopy. Biochemistry 2010, 49 (32), 6846–55. [DOI] [PubMed] [Google Scholar]
- 17.Keizers PM; Ubbink M, Paramagnetic tagging for protein structure and dynamics analysis. Prog Nucl Magn Reson Spectrosc 2011, 58 (1–2), 88–96. [DOI] [PubMed] [Google Scholar]
- 18.Nitsche C; Otting G, Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags. Prog Nucl Magn Reson Spectrosc 2017, 98–99, 20–49. [DOI] [PubMed] [Google Scholar]
- 19.Keyhani S; Goldau T; Blumler A; Heckel A; Schwalbe H, Chemo-Enzymatic Synthesis of Position-Specifically Modified RNA for Biophysical Studies including Light Control and NMR Spectroscopy. Angew Chem Int Ed Engl 2018, 57 (37), 12017–12021. [DOI] [PubMed] [Google Scholar]
- 20.Ramos A; Bayer P; Varani G, Determination of the structure of the RNA complex of a double-stranded RNA-binding domain from Drosophila Staufen protein. Biopolymers 1999, 52 (4), 181–196. [DOI] [PubMed] [Google Scholar]
- 21.Ramos A; Varani G, A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J Am Chem Soc 1998, 120 (42), 10992–10993. [Google Scholar]
- 22.Wu ZY; Lee MD; Carruthers TJ; Szabo M; Dennis ML; Swarbrick JD; Graham B; Otting G, New Lanthanide Tag for the Generation of Pseudocontact Shifts in DNA by Site-Specific Ligation to a Phosphorothioate Group. Bioconjugate Chem 2017, 28 (6), 1741–1748. [DOI] [PubMed] [Google Scholar]
- 23.van Gelder CW; Gunderson SI; Jansen EJ; Boelens WC; Polycarpou-Schwarz M; Mattaj IW; van Venrooij WJ, A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J 1993, 12 (13), 5191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai K; Oubridge C; Jessen TH; Li J; Evans PR, Crystal-Structure of the Rna-Binding Domain of the U1 Small Nuclear Ribonucleoprotein-A. Nature 1990, 348 (6301), 515–520. [DOI] [PubMed] [Google Scholar]
- 25.Oubridge C; Ito N; Evans PR; Teo CH; Nagai K, Crystal-Structure at 1.92 Angstrom Resolution of the Rna-Binding Domain of the U1a Spliceosomal Protein Complexed with an Rna Hairpin. Nature 1994, 372 (6505), 432–438. [DOI] [PubMed] [Google Scholar]
- 26.Oubridge C; Ito N; Teo CH; Fearnley I; Nagai K, Crystallisation of RNA-protein complexes. II. The application of protein engineering for crystallisation of the U1A protein-RNA complex. J Mol Biol 1995, 249 (2), 409–23. [DOI] [PubMed] [Google Scholar]
- 27.Ferre-D’Amare AR, Use of the U1A Protein to Facilitate Crystallization and Structure Determination of Large RNAs. Methods Mol Biol 2016, 1320, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opina ACL; Strickland M; Lee YS; Tjandra N; Byrd RA; Swenson RE; Vasalatiy O, Analysis of the isomer ratios of polymethylated-DOTA complexes and the implictions on protein structural studies. Dalton T 2016, 45 (11), 4673–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haussinger D; Huang JR; Grzesiek S, DOTA-M8: An extremely rigid, high-affinity lanthanide chelating tag for PCS NMR spectroscopy. J Am Chem Soc 2009, 131 (41), 14761–7. [DOI] [PubMed] [Google Scholar]
- 30.Gobl C; Resch M; Strickland M; Hartlmuller C; Viertler M; Tjandra N; Madl T, Increasing the Chemical-Shift Dispersion of Unstructured Proteins with a Covalent Lanthanide Shift Reagent. Angew Chem Int Ed 2016, 55 (47), 14847–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strickland M; Stanley AM; Wang GS; Botos I; Schwieters CD; Buchanan SK; Peterkofsky A; Tjandra N, Structure of the NPr:EINNtr Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems. Structure 2016, 24 (12), 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini I; Luchinat C; Parigi G, Magnetic susceptibility in paramagnetic NMR. Prog Nucl Magn Reson Spectrosc 2002, 40 (3), 249–273. [Google Scholar]
- 33.Sherpa C; Rausch JW; Le Grice SF; Hammarskjold ML; Rekosh D, The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 2015, 43 (9), 4676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sathyamoorthy B; Lee J; Kimsey I; Ganser LR; Al-Hashimi H, Development and application of aromatic [(13)C, (1)H] SOFAST-HMQC NMR experiment for nucleic acids. J Biomol NMR 2014, 60 (2–3), 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schanda P; Brutscher B, Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J Am Chem Soc 2005, 127 (22), 8014–5. [DOI] [PubMed] [Google Scholar]
- 36.Bermejo GA; Clore GM; Schwieters CD, Improving NMR Structures of RNA. Structure 2016, 24 (5), 806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickland M; Schwieters CD; Gobl C; Opina ACL; Strub MP; Swenson RE; Vasalatiy O; Tjandra N, Characterizing the magnetic susceptibility tensor of lanthanide-containing polymethylated-DOTA complexes. J Biomol NMR 2016, 66 (2), 125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John M; Park AY; Pintacuda G; Dixon NE; Otting G, Weak alignment of paramagnetic proteins warrants correction for residual CSA effects in measurements of pseudocontact shifts. J Am Chem Soc 2005, 127 (49), 17190–17191. [DOI] [PubMed] [Google Scholar]
- 39.Cornilescu G; Marquardt JL; Ottiger M; Bax A, Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 1998, 120 (27), 6836–6837. [Google Scholar]
- 40.Biekofsky RR; Muskett FW; Schmidt JM; Martin SR; Browne JP; Bayley PM; Feeney J, NMR approaches for monitoring domain orientations in calcium-binding proteins in solution using partial replacement of Ca2+ by Tb3+. FEBS Lett 1999, 460 (3), 519–26. [DOI] [PubMed] [Google Scholar]
- 41.Feeney J; Birdsall B; Bradbury AF; Biekofsky RR; Bayley PM, Calmodulin tagging provides a general method of using lanthanide induced magnetic field orientation to observe residual dipolar couplings in proteins in solution. J Biomol NMR 2001, 21 (1), 41–48. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q; Stelzer AC; Fisher CK; Al-Hashimi HM, Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 2007, 450 (7173), 1263–U14. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q; Sun XY; Watt ED; Al-Hashimi HM, Resolving the motional modes that code for RNA adaptation. Science 2006, 311 (5761), 653–656. [DOI] [PubMed] [Google Scholar]
- 44.Bermejo GA; Strub MP; Ho C; Tjandra N, Determination of the Solution Bound Conformation of an Amino Acid Binding Protein by NMR Paramagnetic Relaxation Enhancement: Use of a Single Flexible Paramagnetic Probe with Improved Estimation of Its Sampling Space. J Am Chem Soc 2009, 131 (27), 9532–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


