Figure 3.
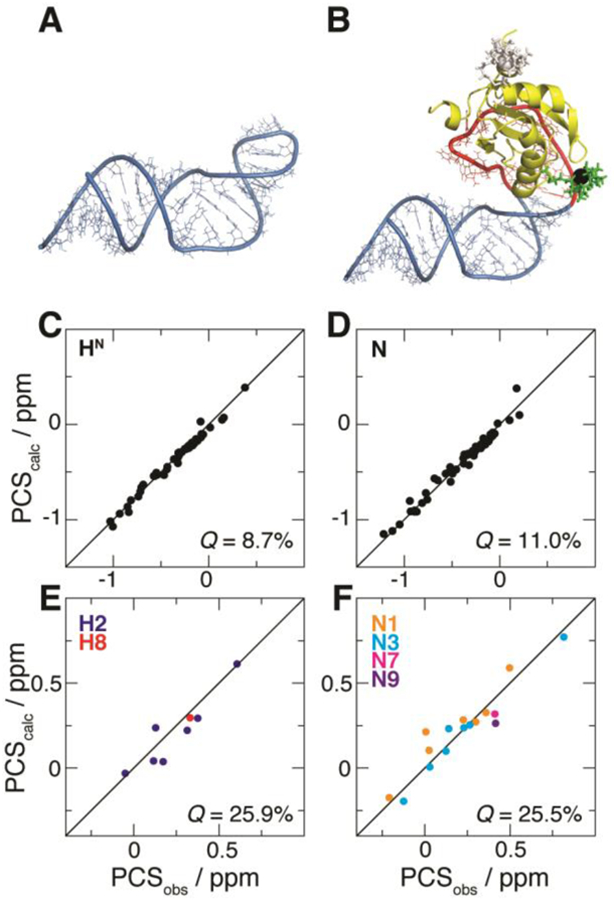
Fit of PCSs from U1A S29C against the structural model of the complex between U1A and the SLCA-based RNA chimera. (A) NMR structure of SLCA.9 (B) Modeled complex between U1A (yellow) and the RNA chimera built with atomic configurations from SLCA (blue) and the 21-nt hairpin (red). The optimized M8-DOTA-SPy conformation is shown (lanthanide, black sphere; remainder, green sticks). For comparison, the conformation of the tag at U1A position 63 (optimized with its corresponding PCSs) is shown (white). (C–F) Correlation plots between observed (PCSobs) and calculated (PCScalc) PCSs for (C) 1H and (D) 15N atoms of U1A (yellow in (B), respectively), and for (E) 1H and (F) 15N atoms of SLCA RNA (blue in (B)). Associated Q-factors are indicated.
