Fig. 3.
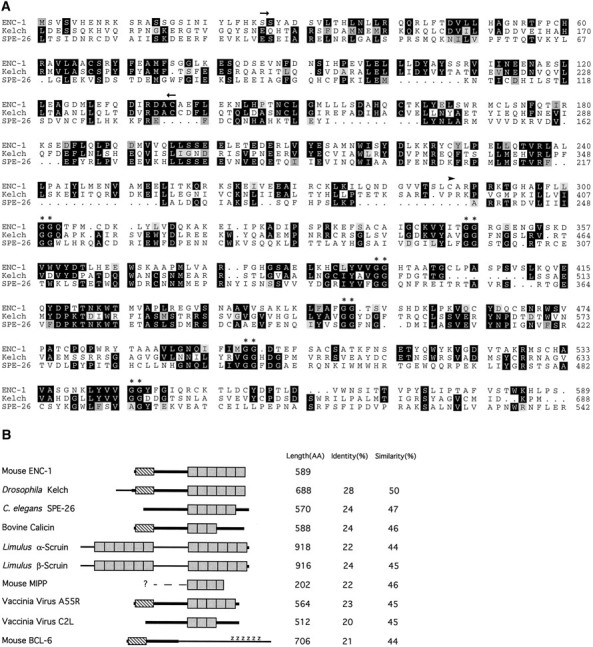
Amino acid comparison of ENC-1 with kelch-related proteins. A, Alignment of ENC-1 and two homologous proteins, kelch and SPE-26, generated by the Wisconsin Genetics Computer Group Sequence Analysis Software Package modules PILE UP and PRETTYBOX. The arrows above the aligned sequences denote a region of homology (BTB/POZ domain) present in ENC-1, kelch, and a number of zinc finger proteins. The arrowhead indicates the start position of the first repeat, based on structural analysis of the repeat motif (kelch motif) by Bork and Doolittle (1994). Thedouble asterisks indicate the double-glycine motif found in all kelch repeats. B, Schematic representation of proteins that share homology to ENC-1. The homology between the proteins is represented by thicker parts oflines representing each sequence. The shaded boxes represent the repeated segments of ∼50 amino acids (kelch motif). The number and location of repeats were based strictly on the presence of two adjacent glycine residues. The hatched box represents the BTB/POZ domain. Z represents the zinc finger regions in the BCL-6 protein. Protein length in amino acids and percentages of amino acid identity and similarity are shown on the right.
