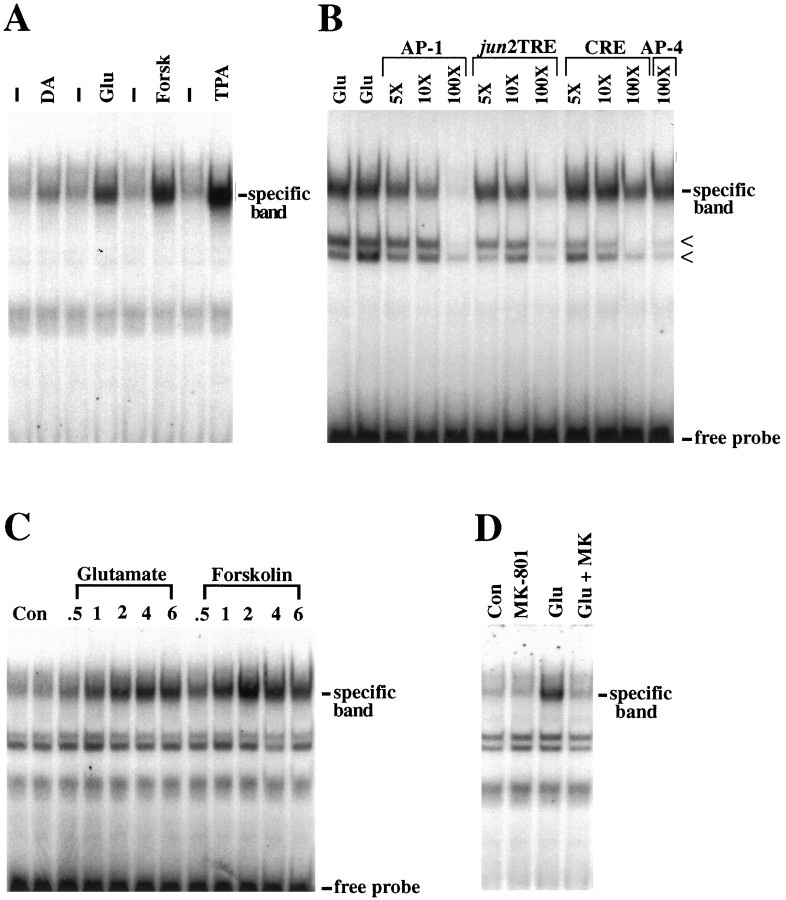
Fig. 2.
AP-1 binding after glutamate, dopamine, and forskolin stimulation in primary striatal cultures. A, AP-1 binding in striatal extracts after stimulation with dopamine (DA, 100 μm), glutamate (Glu, 100 μm), forskolin (Forsk, 10 μm), and TPA (100 nm) for 2 hr. Controls (dash) are from untreated cells. B, The specificity of AP-1-binding activity was determined by gel-shift competition experiments with striatal extracts 2 hr after glutamate stimulation. Unlabeled AP-1 (self), the related oligonucleotides for jun2TRE and CRE, and the unrelated AP-4 element display decreasing potencies, respectively, in their abilities to compete for the upper binding complex (specific band). In addition to the specific band, two less-specific, faster-migrating complexes can be observed (arrowheads). These binding complexes are present in untreated cell extracts and are not glutamate-inducible (see Fig. 3). These complexes are not competed effectively by addition of unlabeled self but can be competed by all the oligonucleotides, including the unrelated AP-4 element, at 100-fold molar excess. C, Time courses of glutamate- and forskolin-induced AP-1 protein binding. After glutamate stimulation, AP-1 binding is induced with a peak at 2–4 hr. Forskolin induction of AP-1 binding is more robust and peaks at 2 hr. D, The induction of AP-1 binding by glutamate stimulation can be blocked by the NMDA receptor antagonist MK-801 (1 μm, added 10 min before glutamate). All data are representative of at least three independent experiments. All experiments were performed with the SP AP-1 oligonucleotide probe.