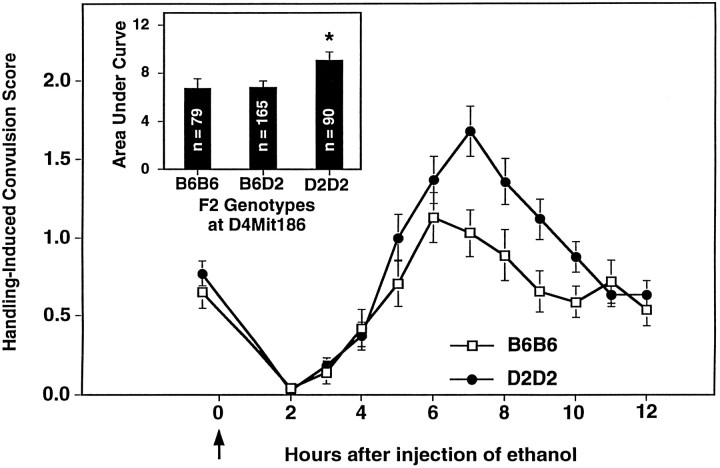
Fig. 2.
Linkage analysis using B6D2 F2 intercross mice provides evidence for a QTL influencing alcohol withdrawal on chromosome 4. Alcohol withdrawal was indexed using the handling-induced convulsion. The mice were scored for baseline handling-induced convulsions immediately before administration of 4 gm/kg ethanol (thearrow marks ethanol injection at time 0), and hourly between 2 and 12 hr after alcohol administration. Data represent the mean raw scores ± SEM for baseline and postethanol handling-induced convulsions. Alcohol administration initially lowers convulsion scores (0–4 hr). Later, convulsion scores increase above baseline, indicating a state of withdrawal hyperexcitability, which peaks ∼6–7 hr after alcohol administration. From 451 F2 intercross mice tested for physiological dependence, we genotyped 167 mice with the highest withdrawal scores and 167 mice with the lowest withdrawal scores. B6D2 F2 mice homozygous for the D2 allele atD4Mit186, a marker located 42.6–45.5 cM from the centromere, showed more severe withdrawal than B6B6 homozygous F2 mice.Inset, Gene dosage at D4Mit186 has a significant influence on alcohol withdrawal severity calculated as area under the curve [F(2,331) = 3.3,p = 0.02). *D2D2 homozygotes have more severe withdrawal than B6B6 homozygotes or B6D2 heterozygotes (Tukey HSD test,p < 0.05).