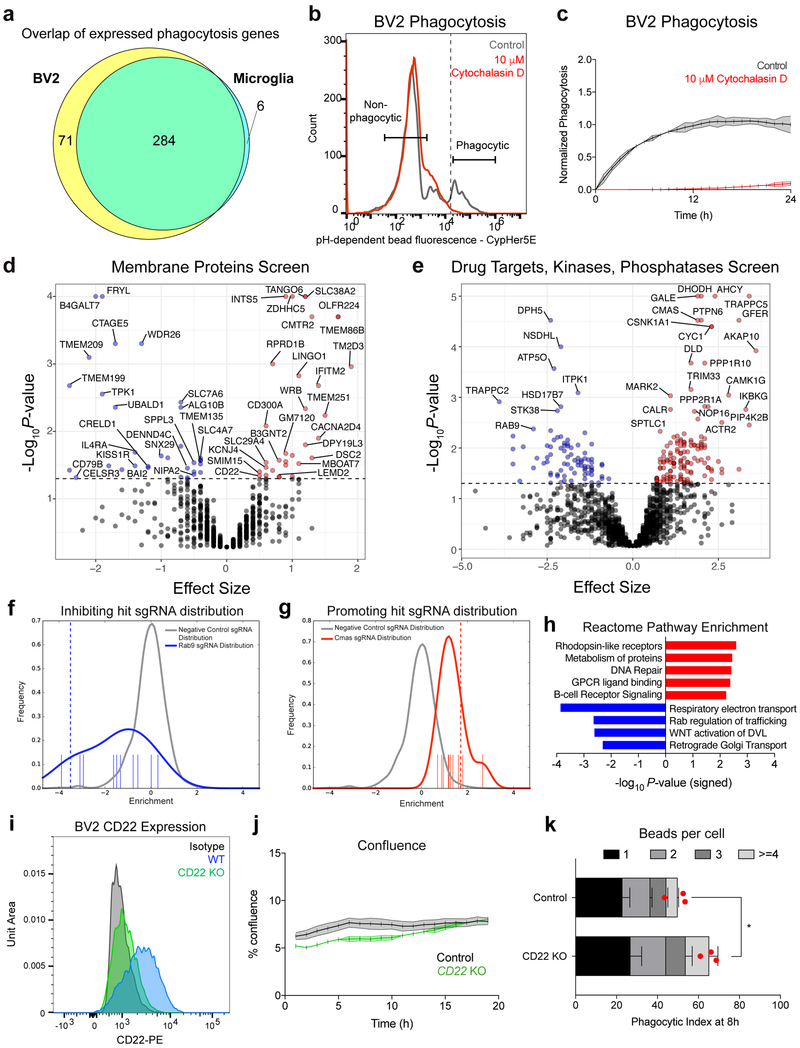
Extended Data Figure 1. CRISPR screen for genetic modifiers of phagocytosis.
a, Intersection and disjunction of 361 genes involved in phagocytosis (Haney et al., 2018) expressed (FPKM>5) by BV2 cells (PRJNA407656) and primary microglia.
b, Gating scheme for FACS separation of phagocytic and non-phagocytic BV2 cells, treated with vehicle (gray) or the actin-polymerization inhibitor, cytochalasin D (red).
c, Time-lapse microscopy readout of phagocytosis by BV2 cells treated with vehicle (gray) or cytochalasin D (red) (n=3, mean +/− s.e.m.).
d, e, Results from CRISPR-Cas9 screen targeting 954 membrane proteins (d) or 2,015 drug targets, kinases, and phosphatases (e) in BV2 cells. Knockouts that promote phagocytosis (red) have a positive effect size and knockouts that inhibit phagocytosis (blue) have a negative effect size (screen performed in technical duplicate; dotted line, P=0.05, two-sided t-test).
f, g, Distributions of negative control sgRNAs (gray) and Rab9-targeting (f) or CMAS-targeting (g) sgRNAs (blue). Positive values indicate enrichment in the phagocytic fraction, and negative values indicate enrichment in the non-phagocytic fraction.
h, Statistical overrepresentation test showing enrichment of Reactome pathway annotations within phagocytosis-promoting (red) and -inhibiting (blue) hits (Fisher’s exact test).
i, CD22 expression in WT (blue), CD22 KO (green), and isotype control stained (black) BV2 cells assessed by flow cytometry.
j, Percent confluence of control (gray) and CD22 KO (green) BV2 cells during time-lapse microscopy phagocytosis assays (n=3, mean +/− s.e.m.).
k, Number of beads ingested per cell were calculated in control and CD22 KO BV2 cells after 8 hours of phagocytosis. While CD22 KO cells display enhanced phagocytosis at a population level (n=3, *P<0.05, two-sided t-test, mean +/− s.e.m.), we observed no significant differences in the number of beads ingested per cell (two-way ANOVA). Red dot represents mean phagocytic index of the entire cell population.
Data in b, c, i, j, k were replicated in at least 2 independent experiments.