Abstract
Kainic acid, the flagship member of the kainoid family of natural neurochemicals, is a widely used neuro-pharmacological agent that helped unravel the key role of ionotropic glutamate receptors, including the kainate receptor, in the central nervous system. Worldwide shortages of this seaweed natural product in the year 2000 prompted numerous chemical syntheses, including scalable preparations with as few as six-steps. Herein we report the discovery and characterization of the concise two-enzyme biosynthetic pathway to kainic acid from l-glutamic acid and dimethylallyl pyrophosphate in red macroalgae and show that the biosynthetic genes are co-clustered in genomes of Digenea simplex and Palmaria palmata. Moreover, we applied a key biosynthetic α-ketoglutarate-dependent dioxygenase enzyme in a biotransformation methodology to efficiently construct kainic acid on the gram scale. This study establishes both the feasibility of mining seaweed genomes for their biotechnological prowess.
Keywords: biosynthesis, biotransformation, kainic acid, natural product, red algae
Graphical Abstract
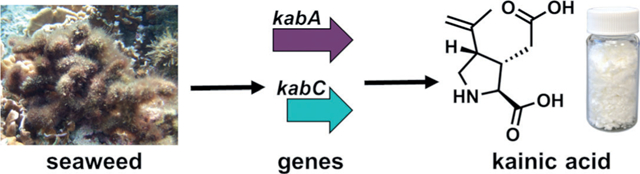
Kainic acid is a marine natural product that has been used as an anthelmintic agent for centuries and, more recently, a neurological tool. Using a genomic sequencing approach, the genes responsible for the biosynthesis of kainic acid were discovered in two different seaweeds. In vitro characterization validated enzymatic activity and enabled gram-scale production of kainic acid using a biotransformation approach.
The tropical seaweed Digenea simplex has been used for centuries in Asia as an anthelmintic agent to treat parasitic worm infections.[1] In the 1950s, the active compound, kainic acid, was isolated,[2,3] enabling its use as a combination treatment for Ascaris infections up until the 1990s.[4–6] While the exact anthelmintic mechanism of action remains unclear, kainic acid functions as an ionotropic glutamate receptor (iGluR) agonist.[7] iGluRs mediate neuronal cell–cell communication by binding to glutamate and facilitating influx of Na+, K+, or Ca2+ into the cell.[8] Structurally similar to glutamic acid, kainic acid can bind more efficiently to iGluRs and thereby stimulate excessive influx of these cations, which leads to excitotoxicity and cell death.[9] In fact, kainic acid was important in the initial discovery and characterization of several classes of iGluRs, such as the eponymous kainate receptor,[10] and has been exploited to create mouse model systems to study neurological diseases,[11] particularly temporal lobe epilepsy.[12,13]
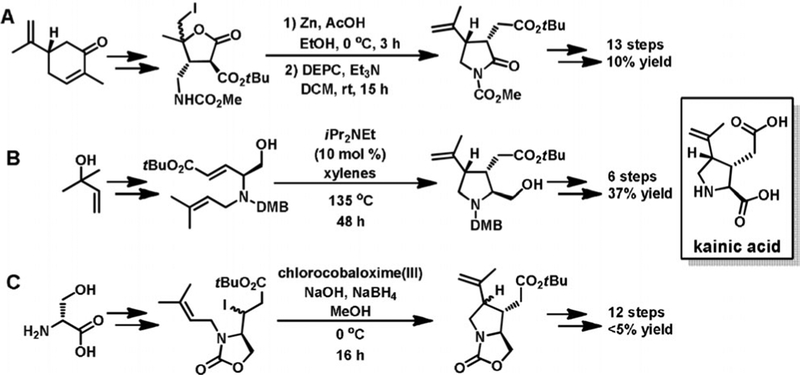
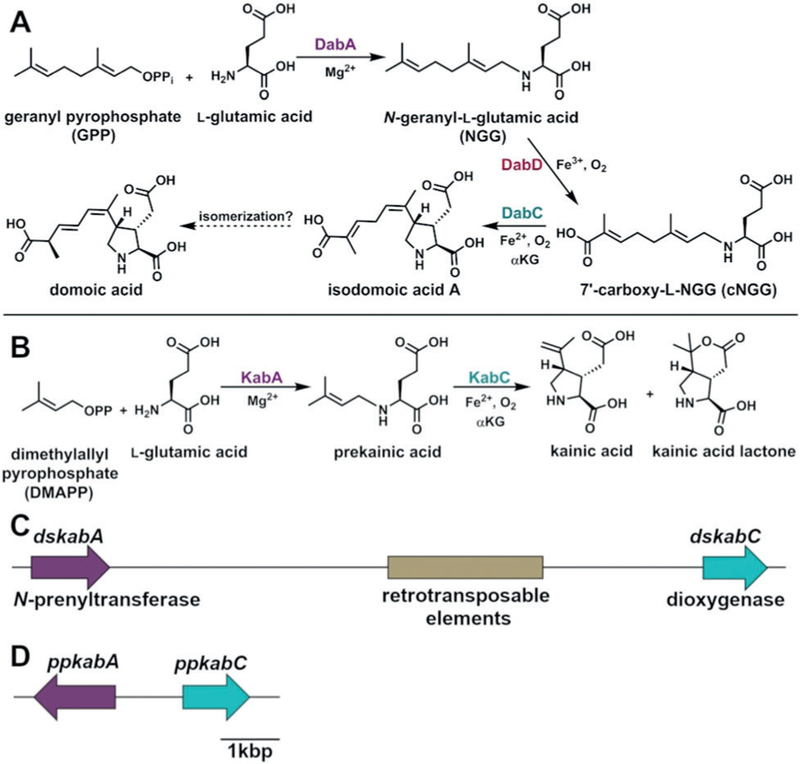
In the 50 years since kainic acid was isolated, over 70 synthetic routes have been established (Figure 1).[14] These syntheses employ diverse methods to produce the kainoid ring pharmacophore such as ring opening followed by cyclization,[15] ene-cyclization,[16] or radical formation.[17] Unfortunately, challenges in generating the three contiguous stereocenters often limit these approaches to low yields or many steps. In contrast to the synthetic work, little progress has been made on elucidating how kainic acid is constructed by seaweeds. Recently, we established the biosynthetic logic for domoic acid production in microalgal Pseudo-nitzschia multiseries diatoms through the discovery of a four-gene cassette (dabA–D) and the confirmation of their in vitro enzymatic functions (Figure 2A).[18] The structural similarity between domoic acid and kainic acid allowed us to propose a conserved route of biosynthesis (Figure 2B). N-prenylation of l-glutamate with dimethylallyl pyrophosphate (DMAPP) via a DabA homolog would produce a prekainic acid pathway intermediate that could conceivably be cyclized directly with a DabC homolog to generate kainic acid.
Figure 1.
Notable synthetic routes to kainic acid by A) Fukuyama,[15] B) Ohshima,[16] and C) Baldwin[17] and co-workers.
Figure 2.
The proposed A) domoic acid[18] and B) kainic acid biosyn-thetic pathways. The kainic acid biosynthesis (kab) gene cluster from C) D. simplex and D) P. palmata. Scale bar is representative for both (C) and (D).
To validate our hypothesis, we performed whole genome sequencing of the well-studied kainic acid producer D. simplex from Japan to identify the kainic acid biosynthetic genes and determine their genetic context. Red macroalgal genome sequencing has proven challenging due to their close association with a diverse array of marine microbes and crustaceans, which means sequencing efforts result in highly complex metagenomes if they are not cured with antibiotics.[19] Moreover, identifying biosynthetic cassettes in red macroalgae genomes, which are often tandem duplications and nested in repetitive sequences, is almost impossible with current high-throughput, short-read (approximately 100 bp) sequencing technologies. However, the new single-molecule, long-read sequencing platforms, such as Oxford Nanopore Technologies (ONT), generate single reads in the hundreds of kilobases (kb), which greatly facilitates resolving tandemly duplicated genes and repeats, assembling metagenomes, and generating highly contiguous reference genomes.[20] We sequenced D. simplex on the ONT MinION platform and produced 7 Gb of sequence with a read N50 of 7 kb and the longest read at 235 kb. We were able to find long reads that contained genes suggestive of kainic acid biosynthesis, but due to the amount of microbial sequence we were not able to assemble the D. simplex genome. Therefore, we sequenced D. simplex on the higher throughput ONT PromethION platform and generated 47 Gb of sequence with a read N50 of 7 kb and the longest read at 1.2 Mb (Supporting Information, Figure S1). The resulting uncorrected genome assembly was 299 Mb with 8246 contigs. The contig N50 was 57 kb and the longest contig was 3.9 Mb. Based on GC% analysis, we predicted that 205 Mb of the assembly was D. simplex, while 88 Mb was high GC% bacterial contigs. This assembly was subsequently polished with Illumina short-read sequencing.
Upon analyzing the D. simplex genome for the presence of homologs to the domoic acid biosynthetic (dab) genes, we uncovered a 12 kb kainic acid biosynthesis (kab) cluster containing an annotated N-prenyltransferase, α-ketoglutarate (αKG)-dependent dioxygenase, and several retrotransposable elements (integrase, reverse transcriptase, and RNase H domains; Figure 2C). Many organisms, including bacteria, fungi, plants, and unicellular algae, are known to cluster genes from the same biosynthetic pathway.[18,21,22] The kab gene clustering is consistent with early examples of red algae clustering genes.[19] To evaluate whether these features are conserved within other kab clusters, we performed genome sequencing of a second known kainic acid producer, Palmaria palmata,[23] to determine the genetic context of its kab genes. Using one MinION chip and no genome assembly steps, we were able to identify a single 47 kb read that contained both ppkabA and ppkabC genes. As with D. simplex, the kainic acid genes are tightly clustered (Figure 2D). However, the gene synteny is not conserved: the gene orientation differs between D. simplex and P. palmata, and a short 1.1 kb intergenic region is located between ppkabA and ppkabC that lacks the retrotransposable elements found in D. simplex.
Analysis of the NCBI transcriptome database identified similar kab transcripts from four different red algae, namely Grateloupia filicina, Palmaria hecatensis, Rhodophysema elegans, and the sequenced P. palmata.[24,25] These three additional red algae are not known kainic acid producers and demonstrate that the genes responsible for biosynthesis are found throughout distinct red algae lineages (Supporting Information, Figure S2). Sequence alignment of the KabA N-prenyltransferase and the KabC αKG-dependent dioxygenase proteins revealed high levels of conservation (Supporting Information, Figures S3 and S4).
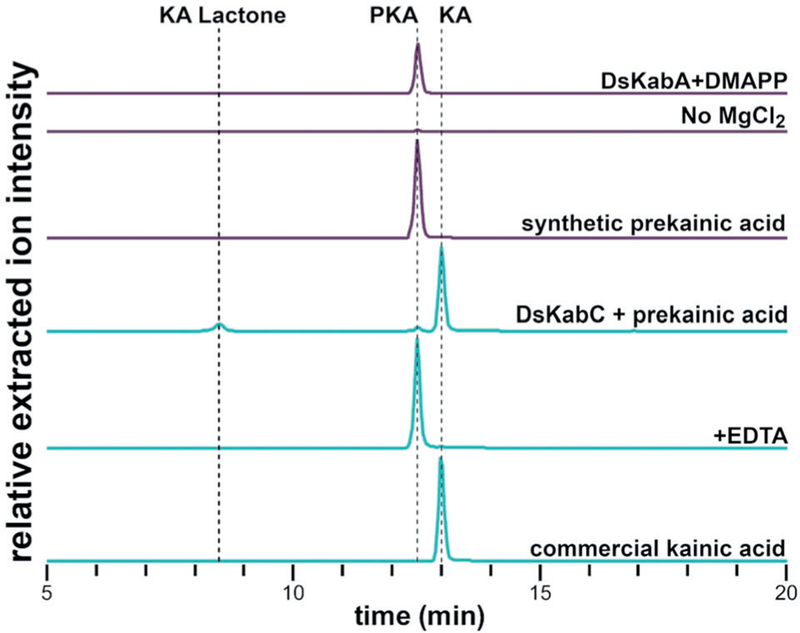
To confirm the proposed kainic acid biosynthetic pathway, dskabA and dskabC were successfully expressed in Escherichia coli and purified to homogeneity (Supporting Information, Figure S5). Incubation of DsKabA with the anticipated substrates DMAPP and l-glutamate produced N-dimethylallyl-l-glutamic acid (prekainic acid) as confirmed by NMR spectroscopy and LCMS comparison with a synthetic standard (Figure 3). Consistent with other members of the terpene cyclase family, Mg2+ is a required cofactor in the reaction. We next explored the substrate selectivity of DsKabA. Exchange of l-glutamate for d-glutamate showed diminished turnover while l-glutamine, l-aspartate, l-asparagine, and glycine did not produce appreciable amounts of N-prenylated products (Supporting Information, Figure S6). In contrast, when we replaced DMAPP with GPP (geranyl pyrophosphate) in the l-glutamate reaction, we clearly observed the formation of N-geranyl-l-glutamic acid (l-NGG), the first biosynthetic precursor of domoic acid (Supporting Information, Figure S6). This high level of amino acid selectivity and modest prenyl donor promiscuity was also observed with DabA from domoic acid biosynthesis.[18] To support the physiological relevance of prekainic acid in the biosynthesis of kainic acid, we examined the aqueous extract of D. simplex. High-resolution LCMS analysis revealed the presence of a compound with both the exact mass and retention time of prekainic acid (Supporting Information, Figure S7).
Figure 3.
LCMS analysis of DsKabA and DsKabC in vitro assays. Each trace represents the (M H)−extracted ion chromatogram for prekainic acid (214.1 ± 0.5 m/z) and kainic acid (212.1 ± 0.5 m/z). DsKabA converts l-Glu and DMAPP to prekainic acid in a Mg2+-dependent manner (purple traces). DsKabC can convert prekainic acid (PKA) to kainic acid (KA) and kainic acid lactone (KA Lactone), but turnover is nearly undetectable when the iron chelator EDTA is added (cyan traces).
After reconstituting DsKabA activity, we next aimed to determine if DsKabC could directly convert prekainic acid to kainic acid. Incubation of DsKabC with prekainic acid, αKG, l-ascorbate, and Fe2+ resulted in the conversion of prekainic acid to two products with the expected mass of kainic acid (Figure 3). Addition of the iron chelator EDTA nearly abolished activity. The major product had the same HPLC retention time and mass as commercially available kainic acid, and was further confirmed by to be kainic acid by NMR spectroscopy. Examination of the in vitro products of three additional algal KabC enzymes revealed a similar trend, wherein prekainic acid is converted to kainic acid and a second compound (Supporting Information, Figure S8). Notably, the KabC ortholog from G. filicinia, GfKabC, produced primarily this other kainic acid isomer. By scaling up the GfKabC reaction, we could isolate this product in sufficient yield to characterize it by NMR spectroscopy, which identified this product as kainic acid lactone (Figure 2B). Kainic acid lactone was previously isolated from kainic-acid-producing red algae[26] and was also found in our D. simplex sample (Supporting Information, Figure S7). In contrast to the agonistic activity of kainic acid, kainic acid lactone has been shown to be an iGluR antagonist.[27] Curiously, the relative ratio of kainic acid to kainic acid lactone appears to vary depending on the KabC ortholog (Supporting Information, Figure S8). While the biochemical basis for this differential product distribution is an ongoing focus of study, we suggest a chemical mechanism analogous to that proposed for DabC[18] that diverges following pyrrolidine ring formation to rationalize the observed chemotype (Supporting Information, Figure S9).
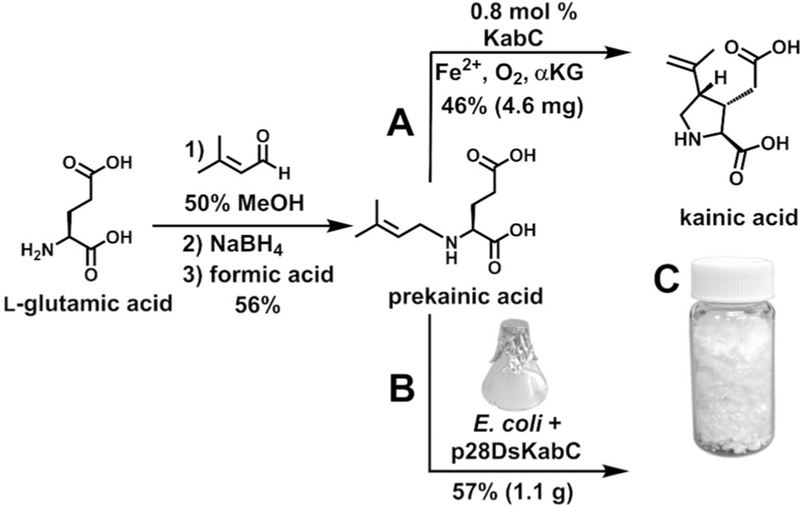
Once the Kab enzymatic functions were validated, we next focused on developing a scalable kainic acid production system that would take advantage of the strengths of both chemical synthesis and biocatalysis. Instead of purifying recombinant KabA and isolating its product from a large-scale biochemical reaction, we readily synthesized prekainic acid by reductive amination of l-glutamate with 3-methyl-2-butenal (Figure 4). The stereospecific cyclization reaction to form the kainic acid pyrrolidine ring, on the other hand, is synthetically challenging and is reminiscent of the cobalt-mediated radical cyclization strategy employed by Baldwin and co-workers in the 1990s on an N-prenylated amino acid substrate (Figure 1C).[17] Therefore, we explored using KabC to produce kainic acid. We first evaluated an in vitro system that used prekainic acid, aKG, Fe2+, and DsKabC, which had been purified from 1 L of E. coli cell culture. After a 16 h incubation, 10 mg of synthetic prekainic acid was completely converted to produce 4.6 mg kainic acid with a 46% isolated yield and a 26% overall isolated yield over two steps (Figure 4A and Supporting Information, Figure S10). Although this chemoenzymatic route was successful, the need to purify DsKabC prevents this method from being applicable for the large-scale production of kainic acid. Therefore, we attempted a biotransformation strategy that would bypass the need to purify enzymes and instead directly convert synthetic prekainic acid to kainic acid in E. coli cells expressing DsKabC (Figure 4B). We developed a streamlined strategy in which prekainic acid was synthesized and the entire unpurified crude reaction mixture was added to the E. coli cell culture. After 40 h, we observed the nearly complete consumption of an 8 mmol prekainic acid synthetic reaction in a biotransformation to kainic acid on a 1 L E. coli cell culture scale (Supporting Information, Figure S11). Employing a two-step purification procedure using activated carbon followed by preparatory reversed phase HPLC, we purified 1.1 g of kainic acid with a combined overall isolated yield of 32% and greater than 95% purity, as assessed by NMR spectroscopy (Figure 4C and Supporting Information, Figure S12).
Figure 4.
Discovery of the kainic acid biosynthetic genes enabled two new kainic acid production routes: A) one-enzyme chemoenzymatic synthesis and B) biotransformation with E. coli cells expressing the dskabC gene in a pET28 vector. C) Lyophilized kainic acid, generated by biotransformation, in a scintillation vial.
Although there are already many scalable synthetic processes for kainic acid, these procedures require at least six synthetic transformations with yields of less than 40% (Figure 1).[14,16] In contrast, biology has evolved an extremely efficient two-step biosynthetic process. The first step, catalyzed by the N-prenyltransferase KabA, is readily replicated using established synthetic procedures. In contrast, the second step, in which KabC catalyzes the stereocontrolled formation of the trisubstituted pyrrolidine ring, is not easily accomplished synthetically. By combining the strengths of prekainic acid synthesis and DsKabC-mediated biocatalysis, we developed a simple and scalable kainic acid production method that is also economically attractive and flexible for analog generation.
Overall, combining genomic information with in vitro enzymatic characterization enabled us to not only discover the enzymes responsible for the biosynthesis of a seaweed natural product, but also to develop new methods to produce this important research tool.
Experimental Section
Experimental details, including DNA sequencing, protein expression, protein purification, production, isolation, chemical synthesis, and spectroscopic characterization are provided in the Supporting Information. All sequence data has been deposited to NCBI databases. The Digenea simplex and Palmaria palmata bioprojects have been deposited as PRJNA509898 and PRJNA509900, respectively. A complete list of all accession numbers can be found in Table S3 in the Supporting Information.
Supplementary Material
Acknowledgements
We thank T. Teruya (University of Ryukyus) for collecting and sending us Digenea simplex, G. Saunders (University of New Brunswick) for sending us DNA and algal samples, T. Steele and E. Ricciardelli (University of California at San Diego) for technical support, and G. Rouse (Scripps Institution of Oceanography) and S. Matsunaga (University of Tokyo) for helpful discussions. This work was supported by the US National Institutes of Health (R01-GM085770 to B.S.M.), the Simons Foundation Fellowship of the Life Science Research Foundation to J.R.C., the Natural Sciences and Engineering Research Council of Canada (NSERC-PDF to S.M.K.M.), and the American Society for Pharmacognosy undergraduate research award to M.L.M.
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201902910.
Conflict of interest
Jonathan R. Chekan, Shaun M. K. McKinnie, and Bradley S. Moore have submitted a provisional patent on the production of kainic acid using the described methods.
Contributor Information
Jonathan R. Chekan, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92093 (USA)
Shaun M. K. McKinnie, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92093 (USA)
Malia L. Moore, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92093 (USA)
Shane G. Poplawski, J. Craig Venter Institute, La Jolla, CA 92037 (USA)
Todd P. Michael, J. Craig Venter Institute, La Jolla, CA 92037 (USA)
Bradley S. Moore, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093 (USA).
References
- [1].Higa T, Kuniyoshi M, Toxicol J. Toxin Rev. 2000, 19, 119–137. [Google Scholar]
- [2].Nitta I, Watase H, Tomiie Y, Nature 1958, 181, 761–762. [DOI] [PubMed] [Google Scholar]
- [3].Murakami S, Takemoto T, Shimizu Z, Yakugaku Zasshi 1953, 73, 1026–1028. [Google Scholar]
- [4].Komiya Y, Kobayashi A, Jpn. J. Med. Sci. Biol 1965, 18, 1–17. [PubMed] [Google Scholar]
- [5].Lee SH, Kang SC, Ahn JH, Lee JW, Rim HJ, Korean J Parasitol. 1972, 10, 79–85. [DOI] [PubMed] [Google Scholar]
- [6].Tremblay J-F, Chem. Eng. News 2000, 78, 14–15. [Google Scholar]
- [7].Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH, Nature 1991, 351, 742–744. [DOI] [PubMed] [Google Scholar]
- [8].Dingledine R, Borges K, Bowie D, Traynelis SF, Pharmacol. Rev 1999, 51, 7–61. [PubMed] [Google Scholar]
- [9].Hampson DR, Manalo JL, Nat. Toxins 1998, 6, 153–158. [DOI] [PubMed] [Google Scholar]
- [10].Lodge D, Neuropharmacology 2009, 56, 6–21. [DOI] [PubMed] [Google Scholar]
- [11].Zheng XY, Zhang HL, Luo Q, Zhu J, J. Biomed. Biotechnol 2011, 2011, 457079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ben-Ari Y, Lagowska J, Tremblay E, Le G La Salle Gal, Brain Res. 1979, 163, 176–179. [DOI] [PubMed] [Google Scholar]
- [13].Lévesque M, Avoli M, Neurosci. Biobehav. Rev 2013, 37, 2887–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stathakis CI, Yioti EG, Gallos JK, Eur. J. Org. Chem 2012, 4661–4673. [Google Scholar]
- [15].Takita S, Yokoshima S, Fukuyama T, Org. Lett 2011, 13, 2068–2070. [DOI] [PubMed] [Google Scholar]
- [16].Zhang M, Watanabe K, Tsukamoto M, Shibuya R, Morimoto H, Ohshima T, Chem. Eur. J 2015, 21, 3937–3941. [DOI] [PubMed] [Google Scholar]
- [17].Baldwin JE, Moloney MG, Parsons AF, Tetrahedron 1990, 46, 7263–7282. [Google Scholar]
- [18].Brunson JK, McKinnie SMK, Chekan JR, McCrow JP, Miles ZD, Bertrand EM, Bielinski VA, Luhavaya H, Oborník M, Smith GJ, Hutchins DA, Allen AE, Moore BS, Science 2018, 361, 1356–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brawley SH, Blouin NA, Ficko-Blean E, Wheeler GL, Lohr M, Goodson HV, Jenkins JW, Blaby-Haas CE, Helliwell KE, Chan CX, Marriage TN, Bhattacharya D, Klein AS, Badis Y, Brodie J, Cao Y, Collän J, Dittami SM, Gachon CMM, Green BR, Karpowicz SJ, Kim JW, Kudahl UJ, Lin S, Michel G, Mittag M, Olson BJSC, Pangilinan JL, Peng Y, Qiu H, Shu S, Singer JT, Smith AG, Sprecher BN, Wagner V, Wang W, Wang Z-Y, Yan J, Yarish C, uner-Riek SZ, Zhuang Y, Zou Y, Lindquist EA, Grimwood J, Barry KW, Rokhsar DS, Schmutz J, Stiller JW, Grossman AR, Prochnik SE, Proc. Natl. Acad. Sci. USA 2017, 114, E6361–E6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Michael TP, Jupe F, Bemm F, Motley ST, Sandoval JP, Lanz C, Loudet O, Weigel D, Ecker JR, Nat. Commun 2018, 9, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boycheva S, Daviet L, Wolfender J, Fitzpatrick TB, Trends Plant Sci. 2014, 19, 447–459. [DOI] [PubMed] [Google Scholar]
- [22].Schläpfer P, Zhang P, Wang C, Kim T, Banf M, Chae L, Dreher K, Chavali AK, Nilo-Poyanco R, Bernard T, Kahn D, Rhee SY, Plant Physiol. 2017, 173, 2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ramsey UP, Bird CJ, Shacklock PF, Laycock MV, Wright JLC, Nat. Toxins 1994, 2, 286–292. [DOI] [PubMed] [Google Scholar]
- [24].Matasci N, Hung L, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, Barker M, Burleigh JG, Gitzendanner MA, Wafula E, Der JP, DePamphilis CW, Roure B, Philippe H, Ruhfel BR, Miles NW, Graham SW, Mathews S, Surek B, Melkonian M, Soltis DE, Soltis PS, Rothfels C, Pokorny L, Shaw JA, DeGironimo L, Stevenson DW, Villarreal JC, Chen T, Kutchan TM, Rolf M, Baucom RS, Deyholos MK, Samudrala R, Tian Z, Wu X, Sun X, Zhang Y, Wang J, Leebens-Mack J, Wong GK-S, Gigascience 2014, 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Saunders GW, Jackson C, Salomaki ED, Mol. Phylogenet. Evol 2018, 119, 151–159. [DOI] [PubMed] [Google Scholar]
- [26].Miyasaki M, Watanabe H, Takano T, Morimoto A, Yakugaku Zasshi 1956, 76, 189–191. [Google Scholar]
- [27].Goldberg O, Luini A, Teichberg VI, Neurosci. Lett 1981, 23, 187–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.