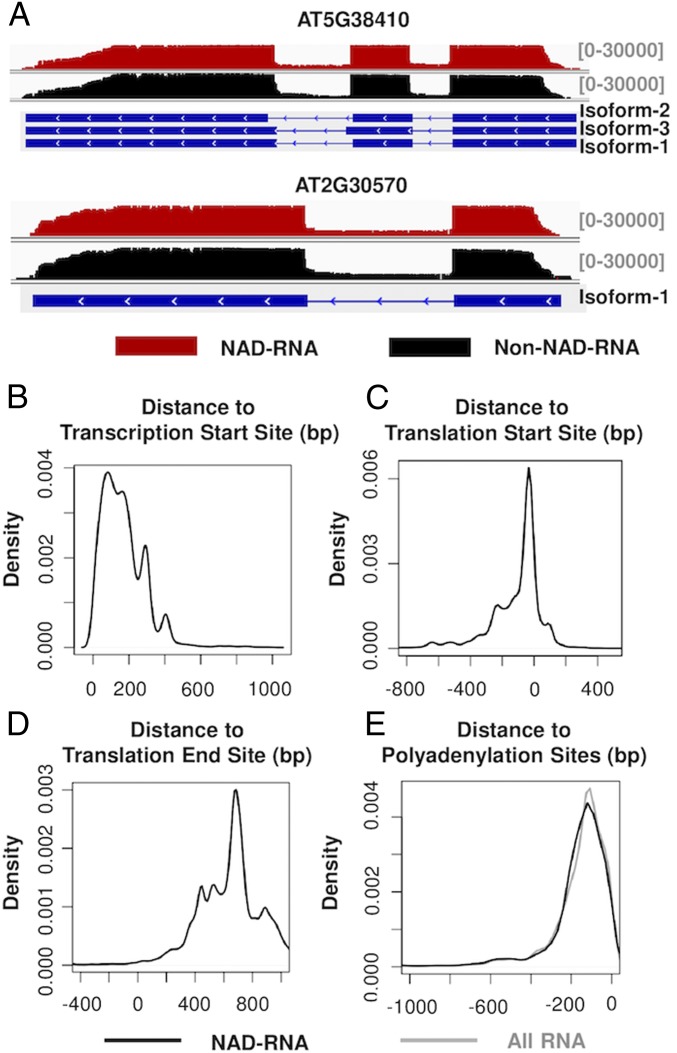
Fig. 5.
Sequence organization of NAD-RNAs. (A) Alignment of NAD-RNA reads and other RNA reads to the genomic sequences of 2 genes. Aligned sequencing reads were visualized using the Integrative Genomics Viewer, with the density of reads denoted above the genes. For the organization of the genes, boxes represent exons, and the lines represent introns. (B) Locations of 5′ ends of the NAD-RNAs relative to annotated TSSs. The reads in the positive x axis had the identified 5′ ends located downstream of the annotated TSSs. (C) Locations of 5′ ends of the NAD-RNAs relative to annotated translation start sites. The reads in the negative x axis had their 5′ ends located upstream of translation start sites. (D) Locations of the 3′ ends of the NAD-RNA reads relative to annotated translation end sites. The reads in the positive x axis contained a 3′ UTR. (E) Distribution of 3′ ends of the NAD-RNA reads (black) and all RNA reads (gray) relative to annotated polyadenylation sites.