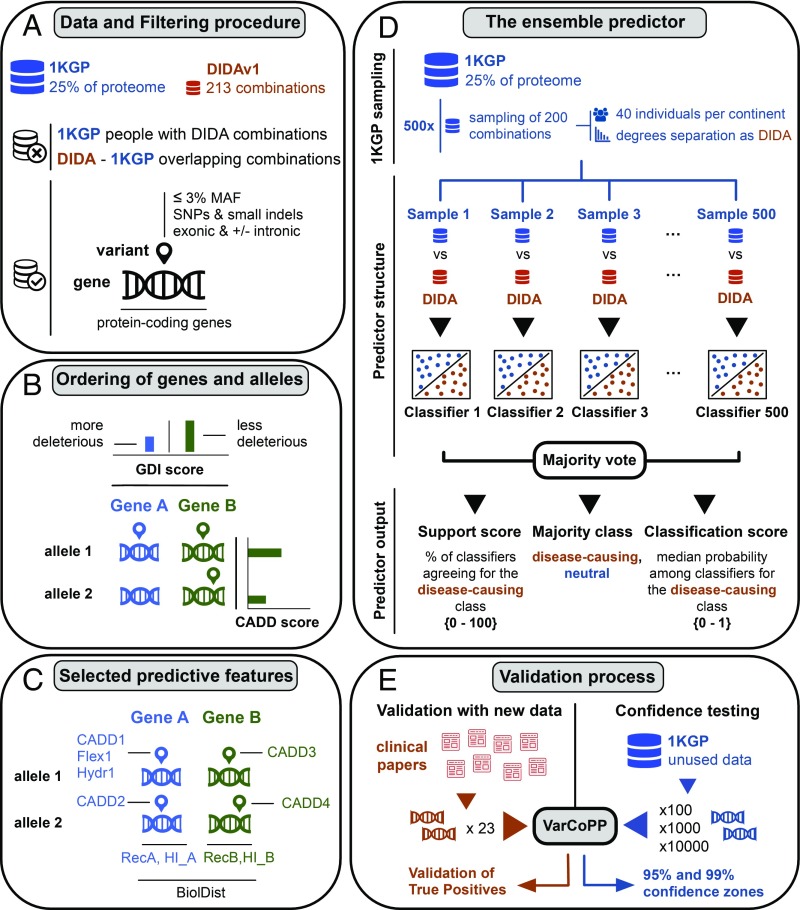
Fig. 3.
Summary of the methodology procedure for the construction of the VarCoPP and the validation process. (A) Genes and variants were filtered in the same way for both the 1KGP and DIDAv1. Individuals of the 1KGP carrying DIDAv1 combinations, as well as the overlapping combinations, were filtered out. Exonic variants [single-nucleotide polymorphism (SNPs) and indels] were used with a MAF frequency of ≤3%, including intronic and synonymous variants close to the exon edges (±13 nucleotides). The genes involved in the procedure were only confirmed protein-coding genes, following the gene types present in the DIDAv1. (B) Bilocus variant combination is represented always using four alleles (two alleles for gene A and two alleles for gene B), including wild-type alleles. This was done in accordance with the information present in the DIDA, where each bilocus combination contained, at maximum, two mutated alleles inside each gene. With this representation, the variant zygosity is also being considered (e.g., for a homozygous variant, both available alleles of the gene contain the same variant information). In this specific panel, we show a bilocus combination with a heterozygous variant in gene A (the second allele is wild-type) and two different heterozygous variants in gene B. Gene A is always the gene with the lowest Gene Damage Index (GDI) score, thus with the higher probability of being a deleterious gene. Different variant alleles inside the same gene were ordered based on their CADD pathogenicity score, with the variant present in the first allele of that gene always having the highest CADD score. (C) Initial number of biological features used for classification was 21, but the final selected and more relevant features were filtered to 11. These included information at the variant level [Flex1 and Hydr1 (i.e., flexibility and hydrophobicity amino acid differences of the first variant allele of gene A), as well as CADD1, CADD2, CADD3, and CADD4, (i.e., the CADD scores of the four different alleles of a bilocus combination)], gene level [RecA, RecB, HI_A, HI_B (i.e., recessiveness and haploinsufficiency probabilities for gene A and gene B)], and gene-pair level [BiolDist (i.e., biological distance, a metric of biological relatedness between two genes of a pair based on protein–protein interaction information)]. A more detailed explanation of the features is provided in SI Appendix, Table S4. (D) After the filtering process, the 1KGP dataset contained billions of bilocus combinations compared with the DIDAv1 set, which contained 200 bilocus combinations. To solve this class imbalance problem, 500 random 1KGP samples, each containing 200 bilocus combinations, were extracted using two types of stratification: Each sample contained an equal amount (41) of bilocus combinations from individuals of each continent as well as an equal distribution of degrees of separation (i.e., a metric of protein–protein interaction distance) between the genes of each pair, following the degrees of separation distribution of the DIDAv1. Each 1KGP sample was used against the complete DIDAv1 set to train an individual classifier that gives a class probability for each bilocus combination. Based on a majority vote among the individual classifiers, the output of the VarCoPP for each tested bilocus combination is the final class (“neutral” or “disease-causing”), the SS (i.e., the percentage of the classifiers agreeing about the pathogenic class), and the CS (i.e., the median probability among the individual predictors that the bilocus combination is pathogenic). (E) To validate the VarCoPP on new disease-causing data, we collected 23 bilocus combinations from independent scientific papers, which included gene pairs not used during the training phase. To perform confidence testing, we extracted three different random sets of 100, 1,000 and 10,000 bilocus combinations from the 1KGP set, which included gene pairs not used during the training phase of the VarCoPP. By exploring the number of FPs predicted with these neutral sets, we defined 95% and 99% confidence zones that provide the minimum SS and CS boundaries above, of which a bilocus combination has a 5% or 1% probability, respectively, of being a FP.