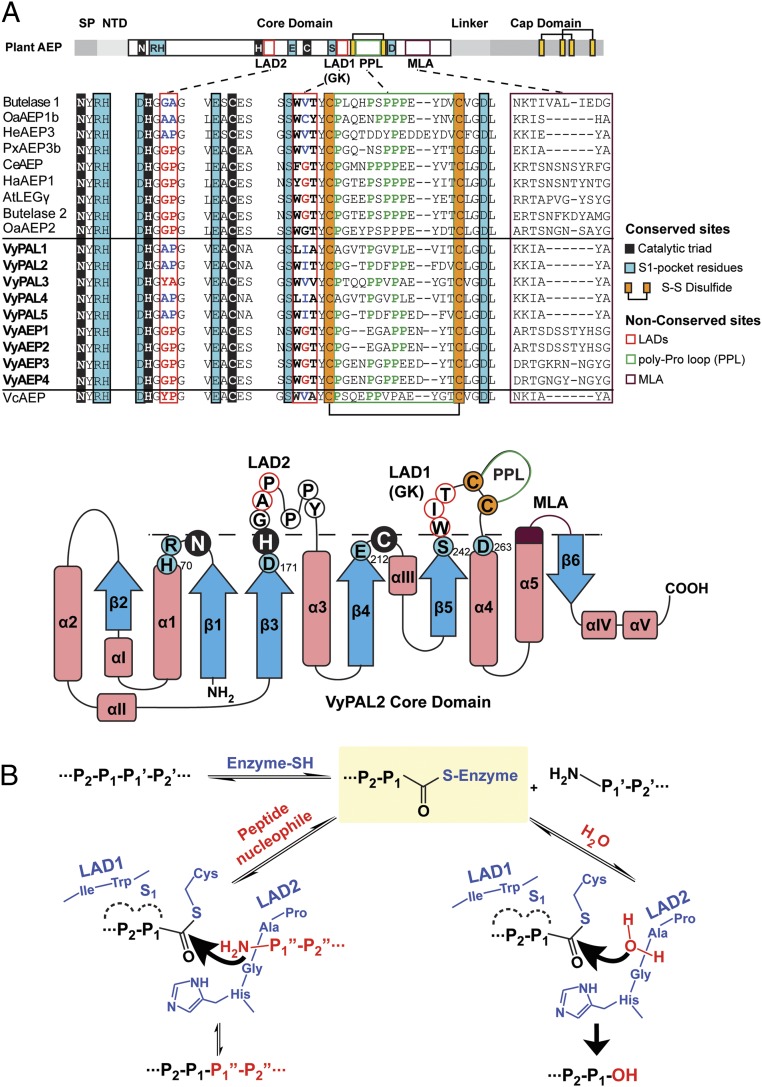
Fig. 6.
Ligase-activity determinants of PALs and the proposed catalytic mechanism. (A) Sequence alignment of PALs and AEPs studied in this work. The catalytic triad Asn–His–Cys is shaded in black. Residues belonging to the S1 pocket are shaded in blue. Proposed LAD residues are boxed in red. Residues of LAD1 and LAD2, that favor ligase activity, are highlighted in blue, and those favoring protease activity are highlighted in red. The conserved disulfide bond near LAD1 is highlighted in orange. The PPL is in a green box and the MLA loop is in purple. The nomenclature of secondary structures was adapted from ref. 48 with alterations according to the crystal structure of VyPAL2 (this work). Residues and motifs crucial for activity are labeled with the same color codes as used in the sequence alignment. Residues below the dashed line correspond to the oxyanion hole, and those above the dashed line correspond to the proposed activity determinants. (B) Schemes proposed for ligation and hydrolysis by VyPAL2 and the role of LAD1 and LAD2. The first step of the mechanism is identical for hydrolysis and ligation, leading to formation of the S-acyl enzyme intermediate, and is the rate-limiting step. Its main determinant is LAD1. LAD2 controls the nature of activity to either favor the nucleophilic attack by a peptide (ligation) or a water molecule (hydrolysis).