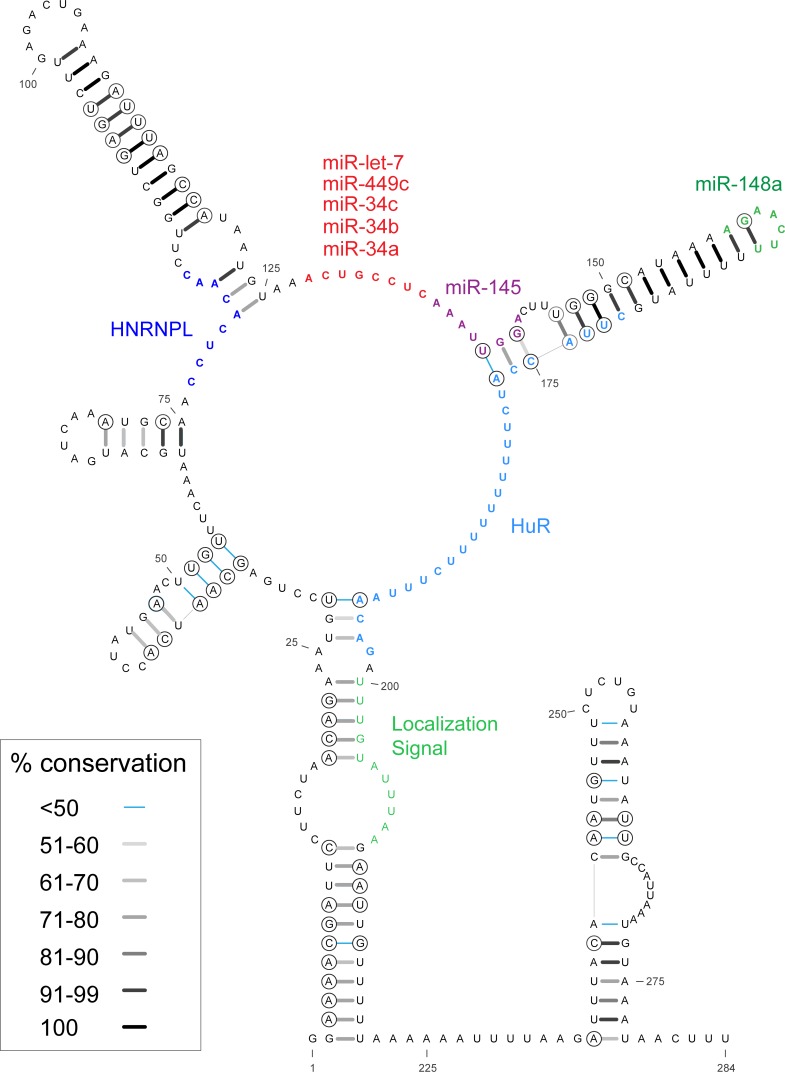
Fig 2. Short MYC 3' UTR model.
Nucleotides 60–177 comprise Motif 17 of the ScanFold-Fold results, which were constrained in the calculation. Base pair conservation shading indicated in the key and data are taken from a comparison of 59 RefSeq mRNA vertebrate alignment (S5 File). Circled bases are sites of putative structure-preserving consistent and compensatory mutations. We have also annotated the sequence with relevant functional motifs: miRNA binding site seed sequences are highlighted (red, purple and green); RBPMap predictions for HNRNPL’s binding sites are colored blue (S7 File; validated by CLIP-seq data [30]); the 28 nt HuR binding site (as reported in [28]) is colored in light blue; and a highly conserved region shown to be a localization signal as reported in [37] is colored in a light green.