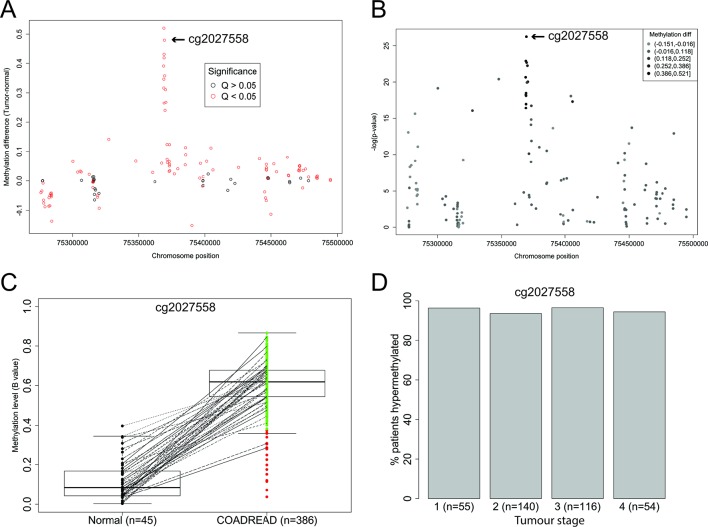
Figure 1.
Levels and prevalence of SEPT9 methylation in colon and rectal adenocarcinoma (COADREAD) compared with adjacent normal tissue in Infinium Human Methylation 450K Array data from The Cancer Genome Atlas (TCGA). (A) Dot plot of differential methylation levels (tumour-normal; y-axis) by genomic location (x-axis) according to the GRCh37/hg19 human genome assembly at each of the SEPT9 probes contained within the 450K array data from the TCGA COADREAD data set. The closest probe to the plasma-based Epi proColon 2.0 CE assay, cg2027558, located at Chr17: 75369484 is indicated. (B) Manhattan plot of the −log10 p values (Wilcoxon rank-sum test; y-axis) for a difference in methylation between tumour and normal tissues by genomic location (x-axis). Probe cg2027558, indicated, showed the most significant difference in tumour versus normal tissue. (C) Spaghetti plot of normalised levels of methylation (β values) at probe cg2027558 in normal tissue and COADREAD tumour tissue. Subject-matched sample pairs are linked by lines. Median, IQRs and 95% CI are indicated. Green=tumours showing hypermethylation, red=tumours with methylation levels in the normal range. (D) Histogram of the frequency of SEPT9 hypermethylation at cg2027558 in colorectal cancer (CRC) from COADREAD stratified by tumour stage.